All
Products
Resources
News
FAQ
Search


Stereo-seq Large Chip Design Solution enables the simultaneous detection of multiple organs within a single section, as well as precisely identifying different cell types within individual organ.
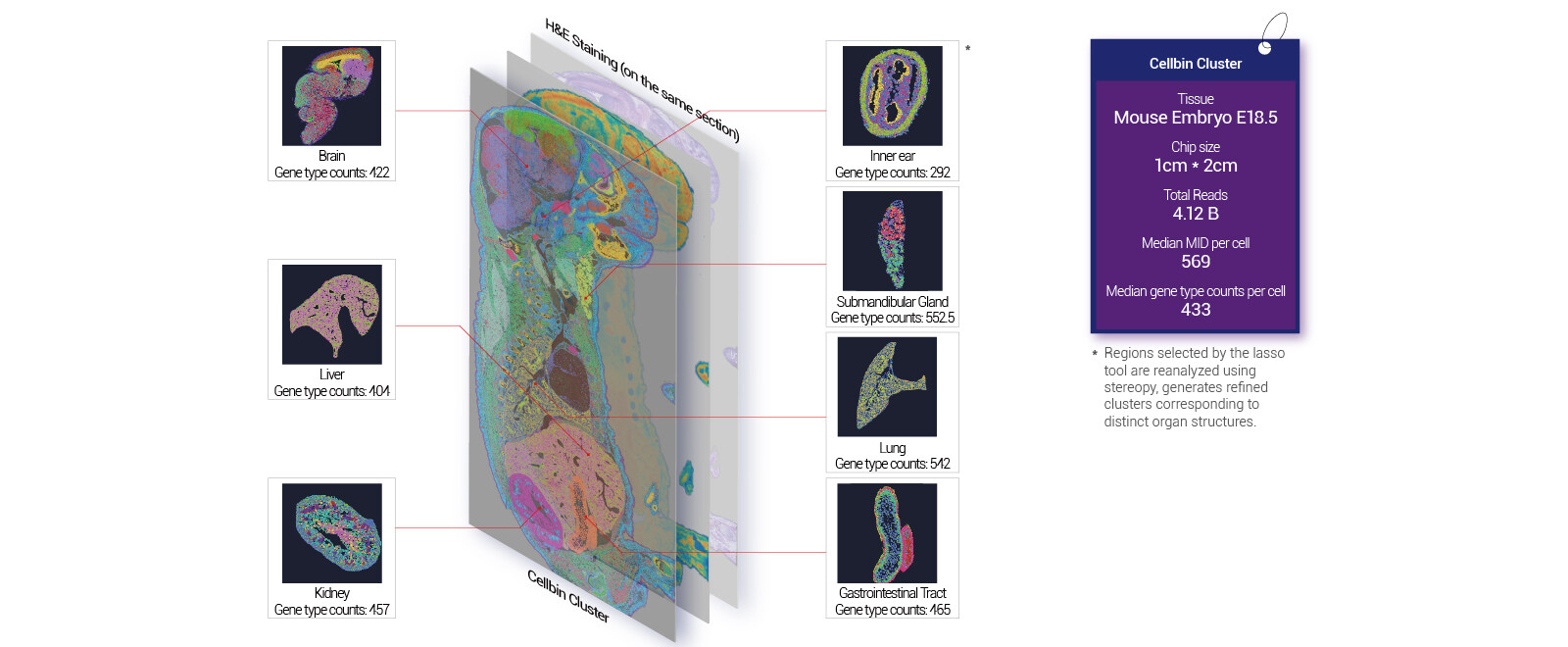
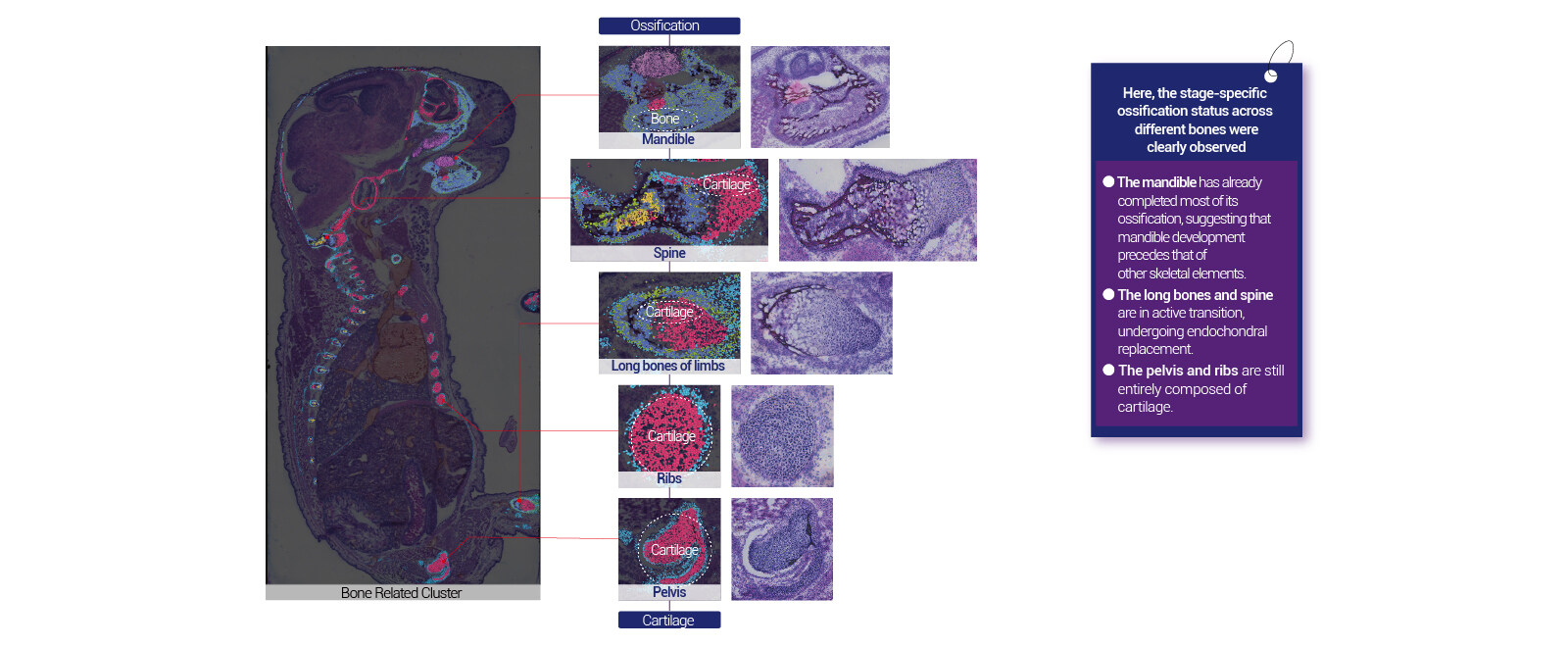
Robust whole-organ and large tissue profiling with single cell resolution

Catalog Number | Catalog Number (US use only) | Set Name | Specifications | Version | Description | |
 | 111SP11124 | 111SP11124-CG | Stereo-seq Permeabilization Set (1cm * 2cm) | 4 RXN | V1.1 | For determining permeabilization parameters to optimize mRNA capture |
111ST13122 | 111ST13122-CG | Stereo-seq Transcriptomics Set (1cm * 2cm) | 2 RXN | V1.3 | For generating a spatially-resolved 3' mRNA library from biological tissue sections | |
 | 111SP11224 | 111SP11224-CG | Stereo-seq Permeabilization Set (2cm * 2cm) | 4 RXN | V1.1 | For determining permeabilization parameters to optimize mRNA capture |
111ST13221 | 111ST13221-CG | Stereo-seq Transcriptomics Set (2cm * 2cm) | 1 RXN | V1.3 | For generating a spatially-resolved 3' mRNA library from biological tissue sections | |
 | 111SP11234 | 111SP11234-CG | Stereo-seq Permeabilization Set (2cm * 3cm) | 4 RXN | V1.1 | For determining permeabilization parameters to optimize mRNA capture |
111ST13231 | 111ST13231-CG | Stereo-seq Transcriptomics Set (2cm * 3cm) | 1 RXN | V1.3 | For generating a spatially-resolved 3' mRNA library from biological tissue sections | |
| 111KL11160 | 111KL11160-CG | Stereo-seq 16 Barcode Library Preparation Kit | 16 RXN | V1.1 | Designed for library preparation of samples using Stereo-seq technology, enables the addition of sample barcodes and library construction. |
Yes, currently only FF V1.3 solution has the LCD format. LCD format for FFPE samples are still in the R&D pipeline and will be released with the next major upgrade of OMNI solution.
The recommended RIN value is derived from R&D tested results. Handling large tissue sections can pose challenges during various experimental steps, such as sample embedding and sectioning. To achieve optimal results, a higher RNA transcript quality standard was recommended for large tissue sections on LCD.
Yes, the DNB libraries of V1.3 different sized chips can be pooled on the same FC for sequencing.