All
Products
Resources
News
FAQ
Search

Analysis tools |
System requirements |
Main function |
Input files |
|---|---|---|---|
SAW |
64-bit Centos/RedHat 78/64 bit Ubuntu 20.04 |
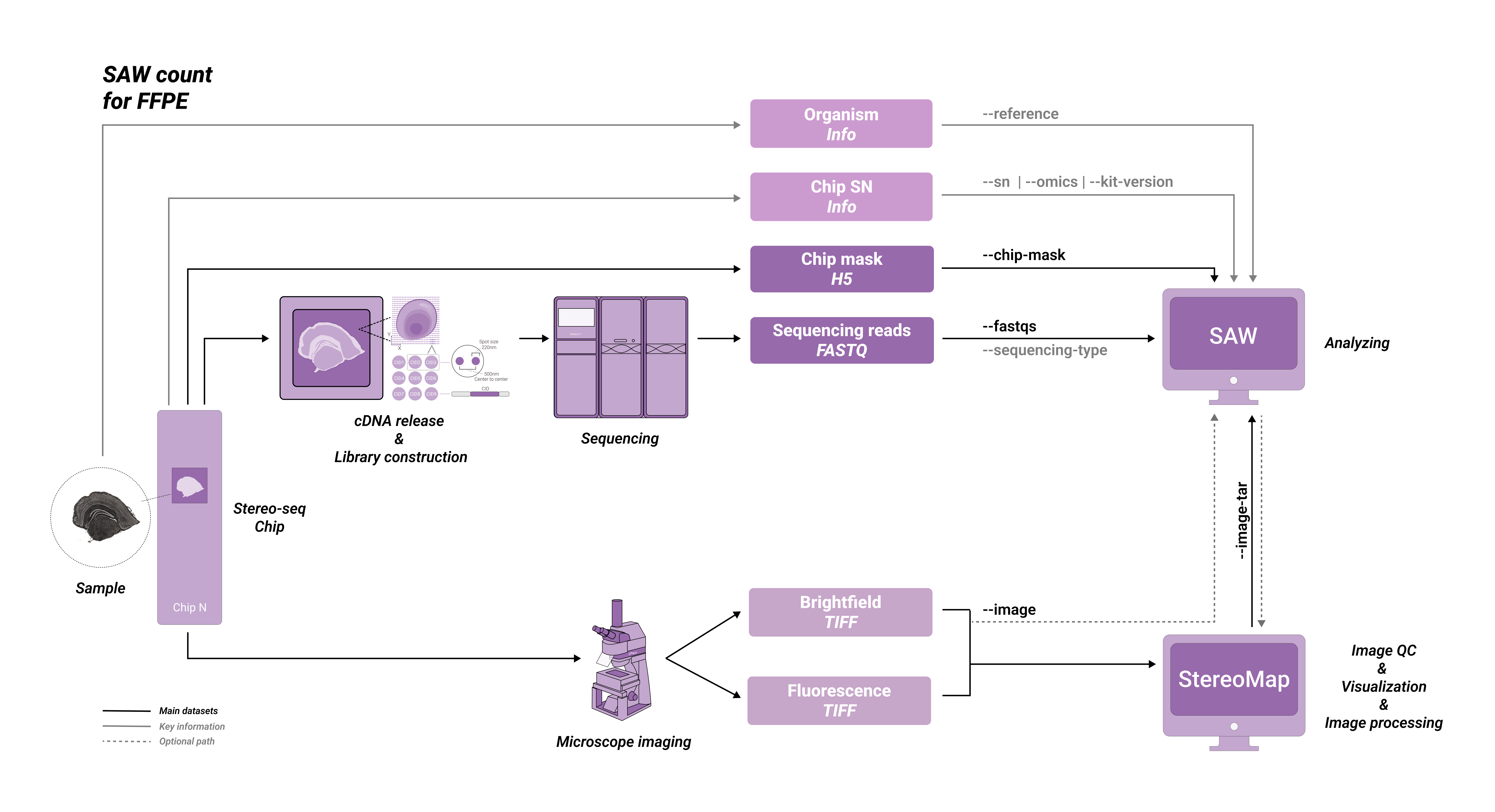
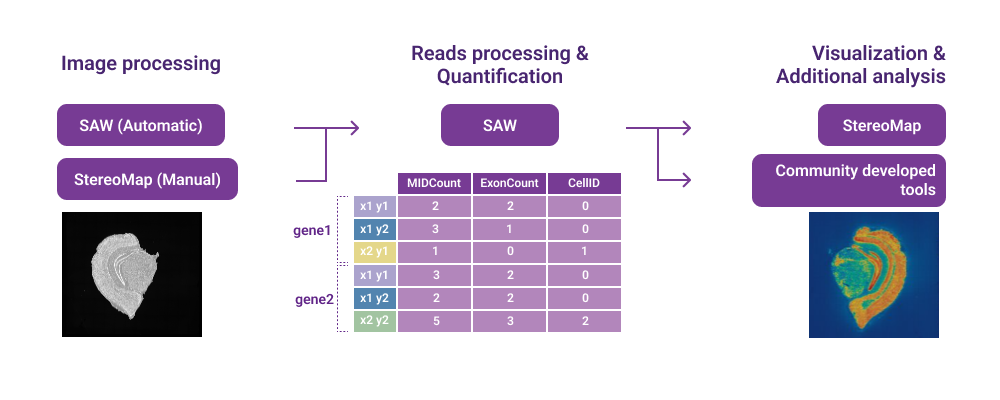
Data filtering, image registration, obtaining of spatial gene expression matrices, and basic secondary analysis. |
References: A reference genome is a comprehensive database of DNA sequences that represents the genomic information of a species. These are available online at databases like Ensembl, UCSC, and NCBI, or you have the option to create your own reference. For microorganism analysis, download index files from Kraken2. Stereo-seq Chip Mask: Stereo-seq chip mask file lists CID (Coordinate ID) sequences for spatial mapping of biological molecules. How to get Mask? Sequencing FASTQ Files: FASTQ files store sequencing data with CID and MID (molecular ID) sequences for spatial identification and molecule quantification. They are generated using DNBSEQ sequencing platforms within your lab or by your DNBSEQ sequencing providers. Images: Microscopic images of tissue sections mounted on Stereo-seq Chips, stained with either nucleus staining or H&E staining, are used to capture tissue morphological information. |
StereoMap |
64-bit Windows 10 & 11 |
Performing visualization and interactive operation on datasets, microscope image quality control, and manual processing step by step. |