All
Products
Resources
News
FAQ
Search
2025.01.09
Paper Review
On January 9, 2025, the STOmics team and MGI Tech Singapore team, in collaboration with clinicians and researchers from Singapore's Duke-NUS Graduate Medical School and the National University of Singapore, published a manuscript titled "Spatially Resolved Tumor Ecosystems and Cell States in Gastric Adenocarcinoma Progression and Evolution." The study, led by Professor Patrick Tan and Dr. Raghav Sundar, was published in Cancer Discovery, a journal of the American Association for Cancer Research (AACR). The team combined whole-ection based Nanostring's GeoMX DSP and paired scRNA-seq to uncover spatially resolved patterns of intratumor heterogeneity in gastric cancer. The STOmics Stereo-seq Fresh Frozen (FF) technology was utilised to increase the spatial resolution beyond Nanostring's GeoMX DSP and to validate the core findings at subcellular resolution. The STOmics experiment was supported by STOmics SG Demo Lab and MGI.
Gastric cancer has a high prevalence in Asia and is the 5th leading cause of cancer mortality worldwide. One of the key challenges in GC treatment is the occurrence of intra-tumoral heterogeneity (ITH), which can occur between subregions in the same tumor, primary and metastatic lesions or even between metastases. This is because ITH can lead to the generation of tumor subclones, facilitating the acquisition of drug resistance. Whilst ITH has been studied at the DNA level (DNA-ITH), investigations into ITH at the transcriptomic level (RNA-ITH) is less explored. The study is groundbreaking as it further incorporates spatial information into the study of RNA-ITH, enabling researchers to understand the interaction between tumor cells and their local tumor microenvironment (TME).
Researchers applied GeoMX DSP and scRNA-seq to analyse 226 samples from 121 patients with GC, integrating data across >2000 regions of interest and studied more than 150,000 single cell transcriptomes. The paired GeoMX DSP / scRNA-seq dataset is one of the largest GC datasets that incorporates both spatial and single-cell level information. This enabled the researchers to identify 2 distinct expression subgroups, which they called “G1” and “G2”. They found that G2 regions were more invasive and are located closer to the tumor edge, whereas the G1 region located in the tumor core. To ensure the reproducibility of their results and to further increase the spatial resolution beyond that achievable by GeoMx DSP, the researchers applied Stereo-seq onto 2 GC samples. They were able to identify the G1 and G2 RNA-ITH subgroups which showed enrichment of pathways similar to those identified with GeoMx DSP.
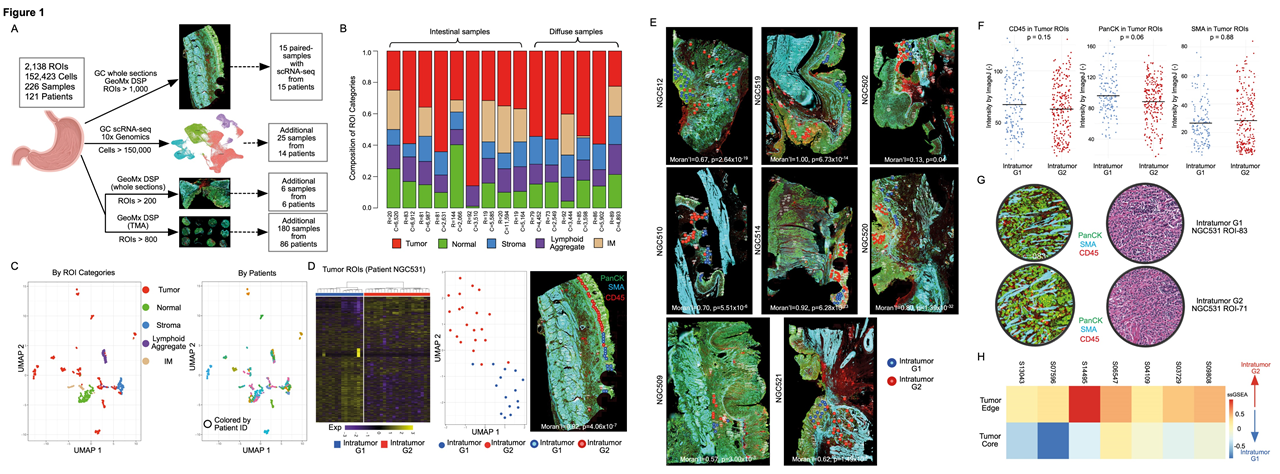
Fig.1 Spatially resolved intratumor heterogeneity (ITH) in gastric cancer
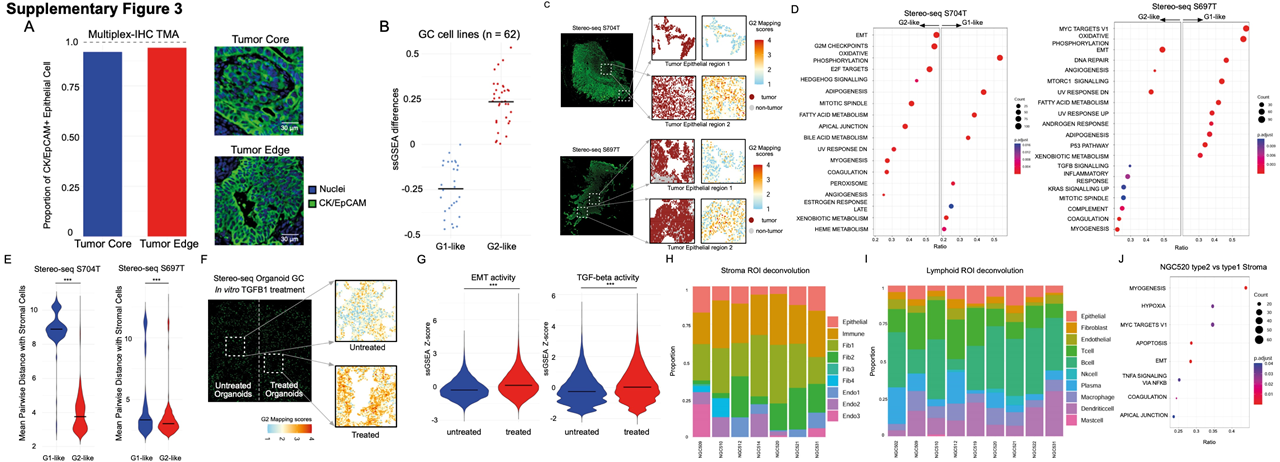
Fig. 2 Stereo-seq was used to evaluate the reproducibility of GeoMx DSP results
Next, the researchers studied the G1 and G2 RNA-ITH subgroups for differences in their local TMEs. They found that the G2 region displayed immune exhaustion signatures, decreased T-cell proportions and elevation of inhibitory cytokines, suggesting that the G2 RNA-ITH regions exhibit an immunosuppressive TME. Interestingly, they also identified potential therapeutic targets such as CAPRIN-1 in the G1 RNA-ITH subregions and Wnt pathway components such as DKK1 and CTNNB1 in the G2 RNA-ITH subregions. This suggests a potential role for spatial RNA-ITH in GC therapy resistance.
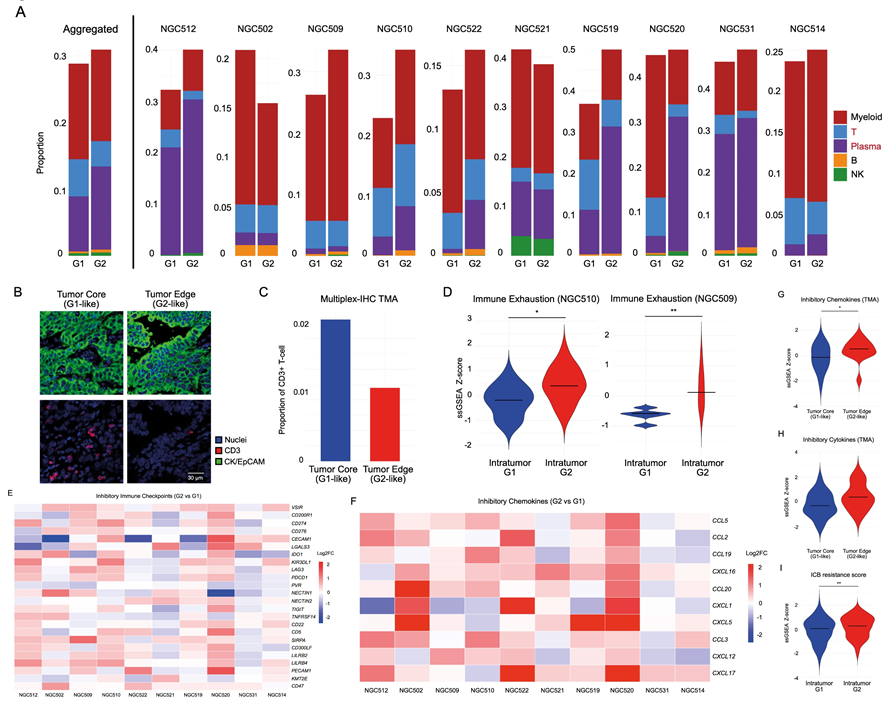
Fig.3 Immune heterogeneity in spatial intratumor subgroups
The authors further examined the relationship between somatic copy number variaitons (sCNAs) and local immune suppression, mediated via the activation of the cGAS-STING pathway. They demonstrated with the GeoMx DSP data that the G2 subregions had higher sCNAs than the G1 subregions. To support their analysis further, they validated their results on the 2 primary GC Stereo-seq samples. They leveraged the popular inferCNV tool and demonstrated that this finding was also recapitulated in the Stereo-seq datasets. As the researchers also collected scRNA-seq data, they further analysed the evolutionary trajectories of the tumor cells and showed that the evolution of tumor subgroups can be classified into 2 models: the branching evolution (gradual accumulation of sCNAs) model and the internal diaspora model (rapid acquisition of sCNAs early in tumor evolution). By comparing their data with The Cancer Genome Atlas gastric cancer (TCGA-STAD) and GASCAD cohorts, they demonstrated that the internal diaspora patient samples have poorer prognosis compared to patients with branched evolution GCs.
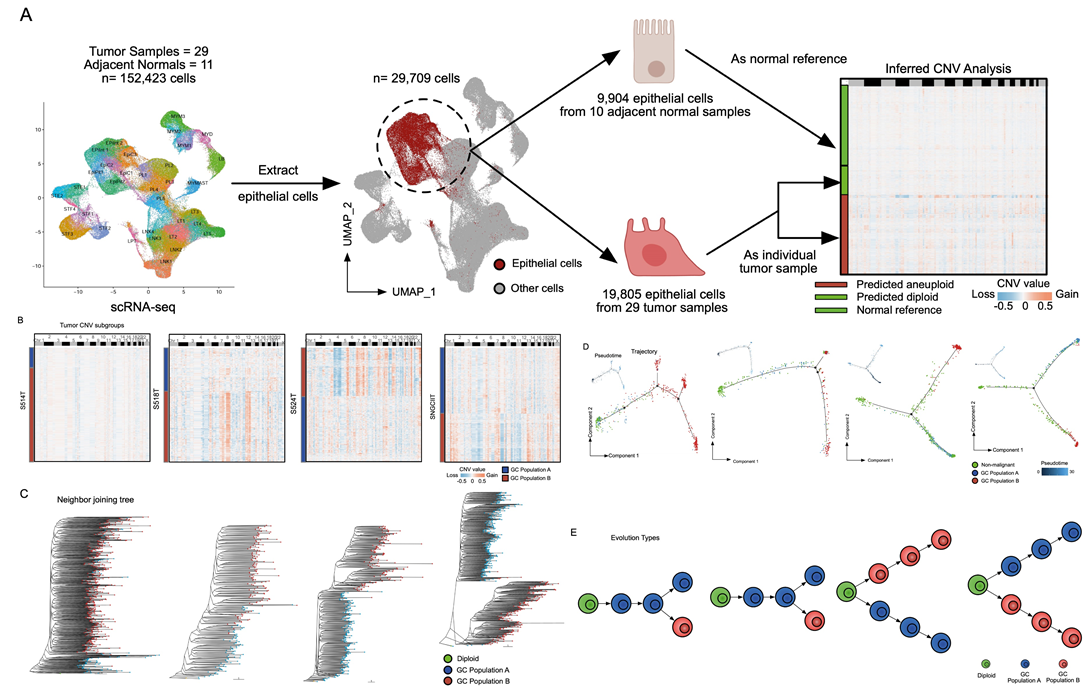
Fig.4 Single-cell resolved evolutionary trajectories in GC.
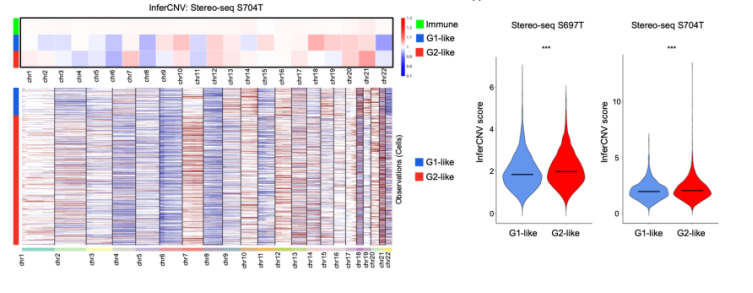
Fig.5 Running InferCNV on Stereo-seq data supports higher sCNAs in G2-like cells.
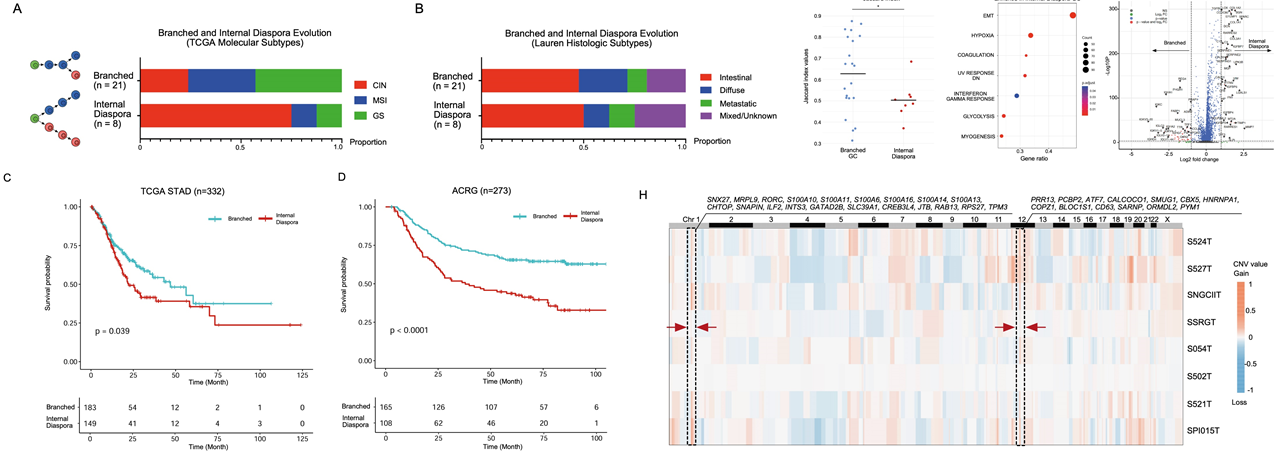
Fig.6 Internal diaspora evolution in GC progression and prognosis.
To study the relationship between the evolutionary patterns (branched evolution vs internal diaspora) and the TME, the researchers investigated the TME patterns in the scRNA-seq dataset and found that internal diaspora GCs have a higher proportion of endothelial cells, fibroblasts and macrophages whereas branched evolution samples have a higher proportion of plasma B cells. They also performed detailed subclustering of these cell types to demonstrate that specific subpopulations such as VWF+ ACKR1+ Endo2 and SPP1+ FN1+ TAM1 (tumor-associated macrophages) are enriched in the internal diaspora GC samples, which are cell types that are known to promote tumor growth and metastases. Their scRNA-seq analyses results are also supported by the GeoMX DSP Stroma ROIs from internal diaspora GCs.
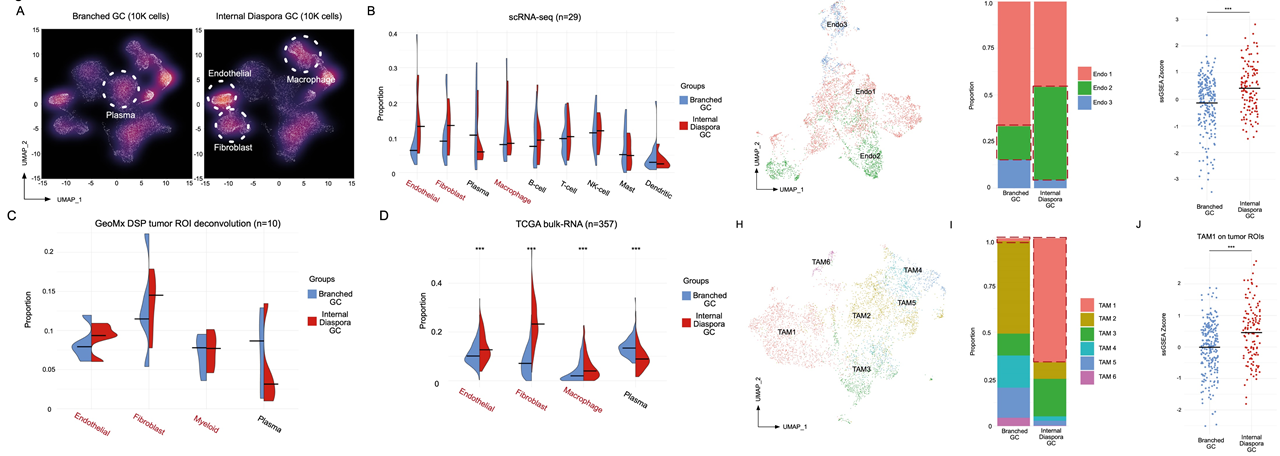
Fig.7 Internal diaspora GC exhibits a unique tumor microenvironment.
The authors then related their analyses of the branched evolution and internal diaspora GCs with the G1-like and G2-like tumor populations. They showed that in the internal diaspora GCs, genes that were upregulated in the G2-like tumor populations such as SOX9, TSPAN8 and AGR2 were also enriched. They further analysed the role of SOX9 by performing an in vitro knockout with the CellOracle tool on a public dataset, as well as in vitro siRNA knockdown and CRISPR-mediated knockout of SOX9 on multiple cell lines. These studies showed that reduction or absence of SOX9 function results in the loss of cell viability and cell migration relative to wild type, suggesting that SOX9 may play a critical role in driving tumor progression in the internal diaspora GCs.
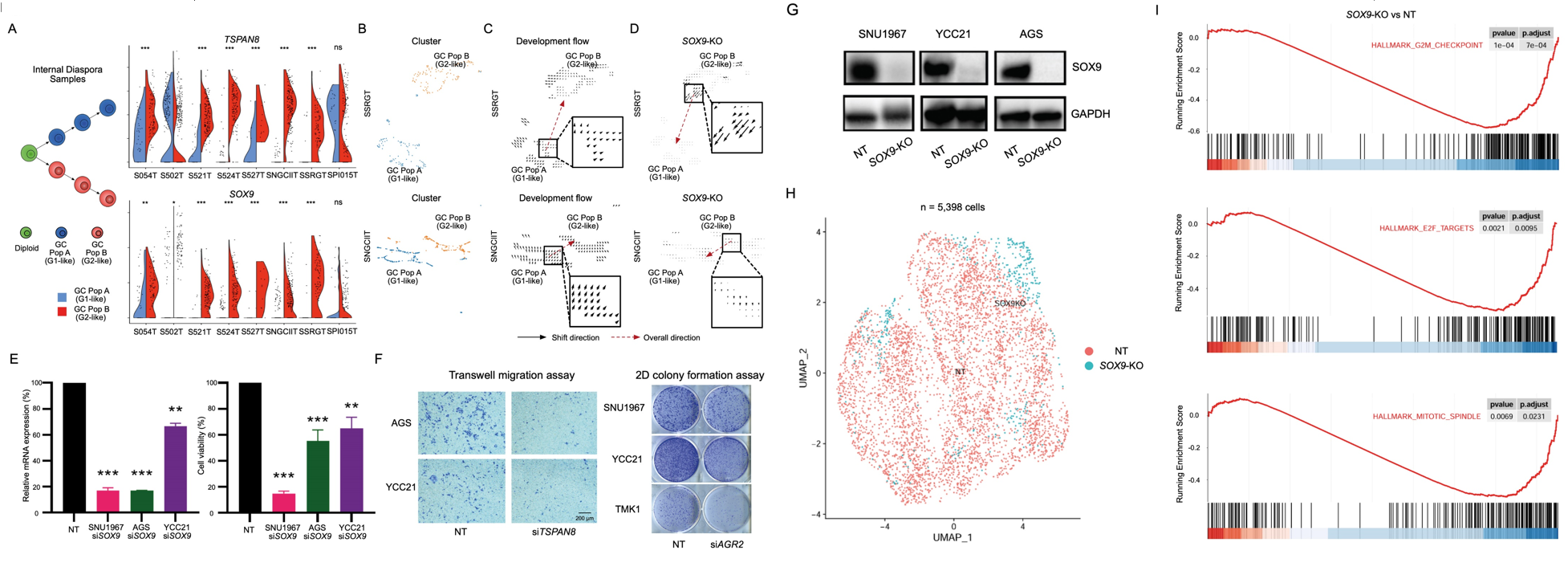
Fig.8 Candidate Drivers of Internal Diaspora Evolution.
Finally, the researchers investigated a specific cancer landmark known as the tumor-stromal interface (TSI). In the TSI, normal and tumor cells are mixed and interact extensively. They found that the GREM1 gene, a gene associated with the TGF-B pathway was enriched in the TSI GeoMx DSP ROIs, which points to the enrichment of myofibroblastic-like cancer-associated fibroblasts (myCAFs). To support this finding, the researchers leveraged the Stereo-seq datasets and were able to distinguish the TSI bins from the stromal and tumor bins via its co-expression of both stromal and tumor markers. By partitioning the tumor regions into two, based on their proximity to stromal cells, the researchers showed that tumor regions closer to stroma cells have upregulated TGF-β levels. Thus, they conclude that GREM1 expression and increased TGF- β signalling may be potential biomarkers of the TSI in GC.
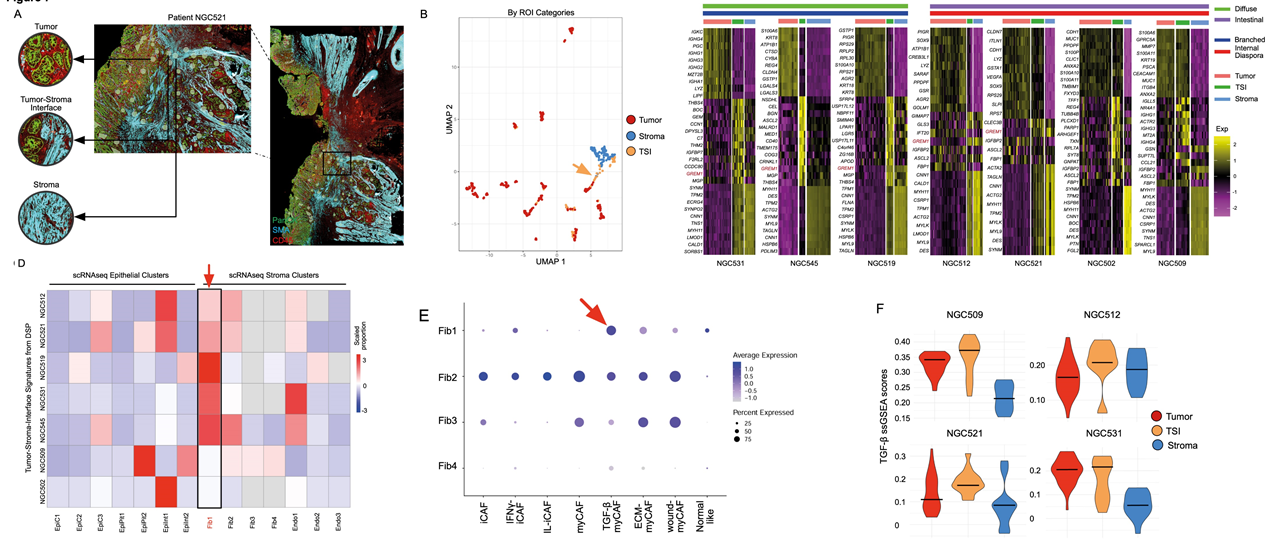
Fig.9 Tumor-stroma interfaces represent a unique TGF-β mediated cell state.
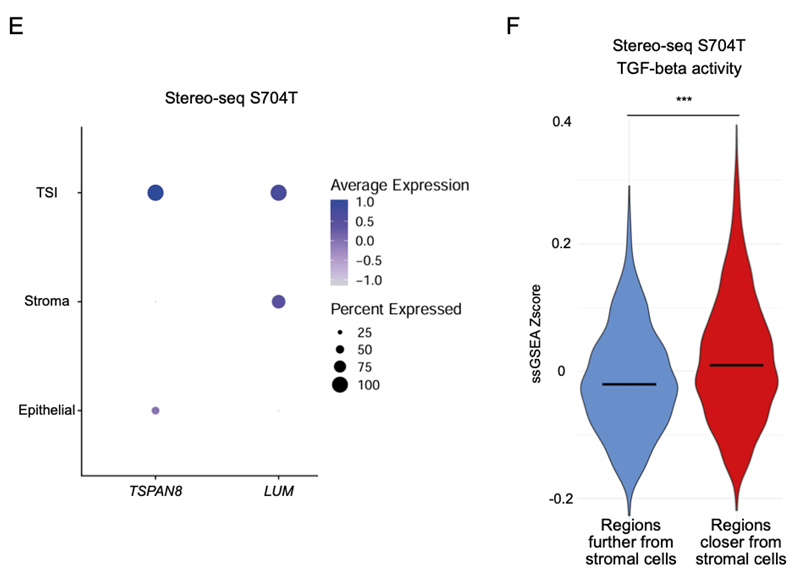
Fig.10 Stereo-seq dataset highlights that regions closer to stromal cells have higher TGF-beta activity.
In conclusion, the researchers created one of the largest GeoMx DSP dataset for GC, supported by orthogonal analyses using Stereo-seq GC data. By combining spatial profiling with single-cell sequencing, they uncovered distinct spatial intratumor subgroups with unique molecular signatures, evolutionary trajectories, and TME characteristics, paving the way towards unveiling potential therapeutic targets for precision medicine approaches for GC.