All
Products
Resources
News
FAQ
Search


$ cat /path/to/output/01.mapping/E100026571_L01_trim_read_1_barcodeMap.stat
...
getBarcodePositionMap_uniqBarcodeTypes: 645784920
total_reads: 1002214171
reads_with_polyA: 131113905 13.08%
reads_filteredByPolyA: 22008148 2.20%
mapped_reads: 826344259 82.45%
reads_with_adapter: 9007116 0.90%
reads_with_dnb: 42264284 4.22%
barcode_exactlyOverlap_reads: 682746301 68.12%
barcode_misOverlap_reads: 143590127 14.33%
barcode_withN_reads: 7831 0.00%
reads_with_rRNA: 0 0.00%
Q10_bases_in_barcode: 99.54%
Q20_bases_in_barcode: 97.49%
Q30_bases_in_barcode: 91.74%
Q10_bases_in_umi: 99.26%
Q20_bases_in_umi: 96.32%
Q30_bases_in_umi: 89.45%
Q10_bases_in_seq: 99.47%
Q20_bases_in_seq: 97.12%
Q30_bases_in_seq: 91.08%
umi_filter_reads: 8265089 0.82%
umi_with_N_reads: 13025 0.00%
umi_with_polyA_reads: 12365 0.00%
umi_with_low_quality_base_reads: 8239699 0.82%
mapped_dnbs: 75619113
...$ cat /path/to/output/01.mapping/E100026571_L01_trim_read_1.Log.final.out
...
Number of input reads | 766807770
Average input read length | 95
UNIQUE READS:
Uniquely mapped reads number | 643871246
Uniquely mapped reads % | 83.97%
Average mapped length | 95.21
Number of splices: Total | 67595584
Number of splices: Annotated (sjdb) | 65674308
Number of splices: GT/AG | 66407685
Number of splices: GC/AG | 457595
Number of splices: AT/AC | 41563
Number of splices: Non-canonical | 688741
Mismatch rate per base, % | 0.50%
Deletion rate per base | 0.07%
Deletion average length | 3.91
Insertion rate per base | 0.03%
Insertion average length | 1.25
MULTI-MAPPING READS:
Number of reads mapped to multiple loci | 87649341
% of reads mapped to multiple loci | 11.43%
Number of reads mapped to too many loci | 5301054
% of reads mapped to too many loci | 0.69%
UNMAPPED READS:
Number of reads unmapped: too many mismatches | 0
% of reads unmapped: too many mismatches | 0.00%
Number of reads unmapped: too short | 28773993
% of reads unmapped: too short | 3.75%
Number of reads unmapped: other | 1212136
% of reads unmapped: other | 0.16%
CHIMERIC READS:
Number of chimeric reads | 0
% of chimeric reads | 0.00% $ samtools view /path/to/output/01.mapping/E100026571_L01_trim_read_1.Aligned.sortedByCoord.out.bam | head -2
E100026571L1C007R00303973559 256 1 3000644 3 100M * 0 0 GCCTCATTGTGCCCCATATGTTTGCCTATGTTGTGGACTTATTTTCATTAAACTTTAAAACATCTTTAATTTTTTTCTTTATTTCATCATTGACCAAGCT -FCA9D?GFFD<-D<cgfegd-dg*fgfdfbe;e(9bgge38fffg9gg;0?ggfgb?e@g:ggg3gf79f0ggdg?g$ head /path/to/output/02.merge/SS200000135TL_D1.merge.barcodeReadsCount.txt
7127 18002 48
4348 19028 1
14130 8635 1
7618 14537 24
4912 10945 5
16783 12914 1
15539 8177 1
9288 8082 14
7274 16533 59
9087 10657 10$ cat /path/to/output/03.count/SS200000135TL_D1.Aligned.sortedByCoord.out.merge.q10.dedup.target.bam.summary.stat
## FILTER & DEDUPLICATION METRICS
TOTAL_READS PASS_FILTER ANNOTATED_READS UNIQUE_READS FAIL_FILTER_RATE FAIL_ANNOTATE_RATE DUPLICATION_RATE
731520587 643871246 532386027 108123310 11.98 17.31 79.69
## ANNOTATION METRICS
TOTAL_READS MAP EXONIC INTRONIC INTERGENIC TRANSCRIPTOME ANTISENSE
643871246 643871246 483163052 49222975 111485219 532386027 109940618
100.0 100.0 75.0 7.6 17.3 82.7 17.1$ samtools view /path/to/output/03.count/SS200000135TL_D1.Aligned.sortedByCoord.out.merge.q10.dedup.target.bam | head -2
E100026571L1C003R03702347721 0 1 3001778 255 100M * 0 0 GTATGACATCTGTCCAGGATCTTCTAGCTTTCATAGTCTCTGGTGAGAAGTCTGGAGTAATTCTAATAGGCCTGCATTTATATGTTACTTGACCTTTTTC EEFEDFFEFFFFEFFFFEC@EFFFFDFFEEFFEFFFFCFCEFFAFBFCED??FGBEFFDC:FFFDCFAF4FAFFDFFDG?DFBD.F@FECA/FEDEFFAA NH:i:1 HI:i:1 AS:i:92 nM:i:3 Cx:i:12136 Cy:i:14034 UR:Z:C0808 XF:i:2
E100026571L1C005R02302788444 528 1 3016331 0 100M * 0 0 TTTATGTGGAGTTCCTTAATCCACTTAGATTTGACCTTAGTACAAGGAGATAGGAATGGATCAATTCGCATTCTTCTACATGATAACAGCCAGTTGTACC ;FDF>FCFFEAD:FFEBF=@FFDEEFFFC@EFCEFDDFFCE?FDFF7EEECFDEFFFCEFCCEEDEEEFEFBFEEFFDEEFFFEEDFFEDFEEEEFFEED NH:i:5 HI:i:1 AS:i:96 nM:i:1 Cx:i:6628 Cy:i:7872 UR:Z:EDFF9$ h5dump -n /path/to/output/03.count/SS200000135TL_D1.raw.gef
HDF5 "/path/to/output/03.count/SS200000135TL_D1.raw.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}
$ h5dump -d /geneExp/bin1/expression /path/to/output/03.count/SS200000135TL_D1.raw.gef | head -15
HDF5 "/path/to/output/03.count/SS200000135TL_D1.raw.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_U32LE "x";
H5T_STD_U32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 76041339 ) / ( 76041339 ) }
DATA {
(0): {
4888,
10392,
1
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/output/03.count/SS200000135TL_D1.raw.gef | head -20
HDF5 "/path/to/output/03.count/SS200000135TL_D1.raw.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 32;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 24661 ) / ( 24661 ) }
DATA {
(0): {
"Gm1992",
0,
132
},
(1): { $ head -8 /path/to/output/03.count/SS200000135TL_D1_raw_barcode_gene_exp.txt
y x geneIndex MIDIndex readCount
10392 4888 10551 665954 4
7096 8901 10551 881671 1
7096 8901 10551 357383 20
18783 7397 10551 355789 1
13032 9155 10551 297666 1
13032 9155 10551 298690 1
11778 10617 10551 686313 4

h5dump -n /path/to/output/04.register/SS200000135TL_D1_20220527_201353_1.1.0.ipr
HDF5 "/path/to/output/04.register/SS200000135TL_D1_20220527_201353_1.1.0.ipr" {
FILE_CONTENTS {
group /
group /DAPI
group /DAPI/CellSeg
dataset /DAPI/CellSeg/CellMask
group /DAPI/ImageInfo
dataset /DAPI/ImageInfo/RGBScale
dataset /DAPI/Preview
group /DAPI/QCInfo
group /DAPI/QCInfo/CrossPoints
dataset /DAPI/QCInfo/CrossPoints/0_0
...
dataset /DAPI/QCInfo/CrossPoints/9_7
dataset /DAPI/QCInfo/ScopeStitchQCMatrix
group /DAPI/Register
dataset /DAPI/Register/MatrixTemplate
group /DAPI/Stitch
group /DAPI/Stitch/BGIStitch
dataset /DAPI/Stitch/BGIStitch/StitchedGlobalLoc
group /DAPI/Stitch/ScopeStitch
dataset /DAPI/Stitch/ScopeStitch/GlobalLoc
group /DAPI/Stitch/StitchEval
dataset /DAPI/Stitch/StitchEval/StitchEvalH
dataset /DAPI/Stitch/StitchEval/StitchEvalV
dataset /DAPI/Stitch/TemplatePoint
dataset /DAPI/Stitch/TransformTemplate
group /DAPI/TissueSeg
dataset /DAPI/TissueSeg/TissueMask
group /ManualState
group /StereoResepSwitch
}
}
$ h5dump -A /path/to/output/04.register/SS200000135TL_D1_20220527_201353_1.1.0.ipr | head -20
HDF5 "/path/to/output/04.register/SS200000135TL_D1_20220527_201353_1.1.0.ipr" {
GROUP "/" {
ATTRIBUTE "IPRVersion" {
DATATYPE H5T_STRING {
STRSIZE H5T_VARIABLE;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_UTF8;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "0.1.0"
}
}
GROUP "ManualState" {
ATTRIBUTE "cellseg" {
DATATYPE H5T_ENUM {
H5T_STD_I8LE;
"FALSE" 0;
"TRUE" 1;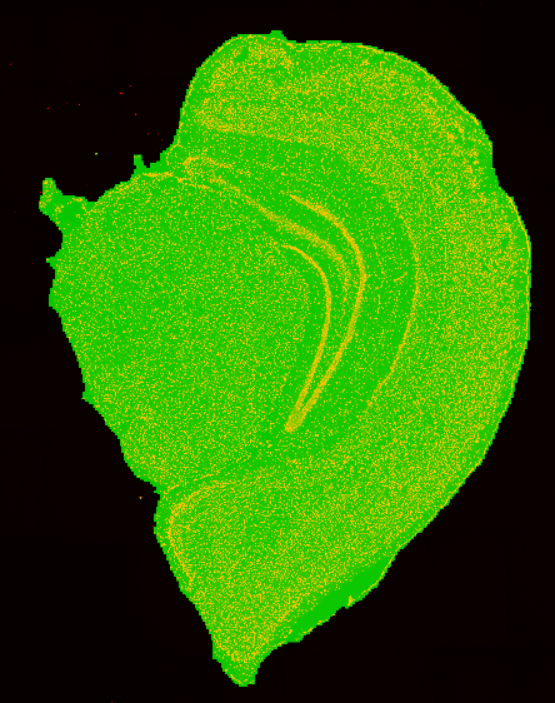

$ cat /path/to/output/05.tissuecut/tissuecut.stat
# Tissue Statistic Analysis with Stain Image
Contour_area: 88648172
Number_of_DNB_under_tissue: 36681107
Ratio: 41.38%
Total_gene_type: 24300
MID_counts: 89818303
Fraction_MID_in_spots_under_tissue: 83.07%
Reads_under_tissue: 648385443
Fraction_reads_in_spots_under_tissue: 78.46%
binSize=1
Mean_reads_per_spot: 17.68
Median_reads_per_spot: 11.00
Mean_gene_type_per_spot: 1.71
Median_gene_type_per_spot: 1
Mean_Umi_per_spot: 2.45
Median_Umi_per_spot: 2
binSize=20
Mean_reads_per_spot: 2911.89
Median_reads_per_spot: 2445.00
Mean_gene_type_per_spot: 237.55
Median_gene_type_per_spot: 223
Mean_Umi_per_spot: 403.37
Median_Umi_per_spot: 364
binSize=50
Mean_reads_per_spot: 18042.28
Median_reads_per_spot: 16195.00
Mean_gene_type_per_spot: 1151.00
Median_gene_type_per_spot: 1117
Mean_Umi_per_spot: 2499.33
Median_Umi_per_spot: 2308
binSize=100
Mean_reads_per_spot: 71102.70
Median_reads_per_spot: 64449.00
Mean_gene_type_per_spot: 3082.76
Median_gene_type_per_spot: 3080
Mean_Umi_per_spot: 9849.58
Median_Umi_per_spot: 9066
binSize=150
Mean_reads_per_spot: 157566.33
Median_reads_per_spot: 143745.00
Mean_gene_type_per_spot: 4890.24
Median_gene_type_per_spot: 5026
Mean_Umi_per_spot: 21827.05
Median_Umi_per_spot: 20205
binSize=200
Mean_reads_per_spot: 276733.00
Median_reads_per_spot: 254272.00
Mean_gene_type_per_spot: 6403.57
Median_gene_type_per_spot: 6719
Mean_Umi_per_spot: 38334.74
Median_Umi_per_spot: 35679 $ h5dump -n /path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
$ h5dump -d /geneExp/bin1/expression /path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef | head -15
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_U32LE "x";
H5T_STD_U32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 62649229 ) / ( 62649229 ) }
DATA {
(0): {
9915,
15992,
1
},
(1): {
h5dump -d /geneExp/bin1/gene /path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef | head -20
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 32;
STRPAD H5T_STR_NULLPAD;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 24300 ) / ( 24300 ) }
DATA {
(0): {
"Gm37381",
0,
3
},
(1): {$ h5dump -n /path/to/output/05.tissuecut/SS200000135TL_D1.gef
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
group /geneExp/bin10
dataset /geneExp/bin10/exon
dataset /geneExp/bin10/expression
dataset /geneExp/bin10/gene
group /geneExp/bin100
dataset /geneExp/bin100/exon
dataset /geneExp/bin100/expression
dataset /geneExp/bin100/gene
group /geneExp/bin20
dataset /geneExp/bin20/exon
dataset /geneExp/bin20/expression
dataset /geneExp/bin20/gene
group /geneExp/bin200
dataset /geneExp/bin200/exon
dataset /geneExp/bin200/expression
dataset /geneExp/bin200/gene
group /geneExp/bin50
dataset /geneExp/bin50/exon
dataset /geneExp/bin50/expression
dataset /geneExp/bin50/gene
group /geneExp/bin500
dataset /geneExp/bin500/exon
dataset /geneExp/bin500/expression
dataset /geneExp/bin500/gene
group /stat
dataset /stat/gene
group /wholeExp
dataset /wholeExp/bin1
dataset /wholeExp/bin10
dataset /wholeExp/bin100
dataset /wholeExp/bin20
dataset /wholeExp/bin200
dataset /wholeExp/bin50
dataset /wholeExp/bin500
group /wholeExpExon
dataset /wholeExpExon/bin1
dataset /wholeExpExon/bin10
dataset /wholeExpExon/bin100
dataset /wholeExpExon/bin20
dataset /wholeExpExon/bin200
dataset /wholeExpExon/bin50
dataset /wholeExpExon/bin500
}
}
$ h5dump -d /stat/gene /path/to/output/05.tissuecut/SS200000135TL_D1.gef | head -20
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.gef" {
DATASET "/stat/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 32;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "MIDcount";
H5T_IEEE_F32LE "E10";
}
DATASPACE SIMPLE { ( 24661 ) / ( 24661 ) }
DATA {
(0): {
"Gm42418",
5861037,
60.1033
},
(1): {
$ h5dump -n /path/to/output/051.cellcut/SS200000135TL_D1.cellbin.gef
HDF5 "/path/to/output/051.cellcut/SS200000135TL_D1.cellbin.gef" {
FILE_CONTENTS {
group /
group /cellBin
dataset /cellBin/blockIndex
dataset /cellBin/blockSize
dataset /cellBin/cell
dataset /cellBin/cellBorder
dataset /cellBin/cellExon
dataset /cellBin/cellExp
dataset /cellBin/cellExpExon
dataset /cellBin/cellTypeList
dataset /cellBin/gene
dataset /cellBin/geneExon
dataset /cellBin/geneExp
dataset /cellBin/geneExpExon
}
} $ cat /path/to/output/07.saturation/sequence_saturation.tsv
sample bar_x bar_y1 bar_y2 bar_umi bin_x bin_y1 bin_y2 bin_umi
0.05 26619302 0.250959 1 19938952 26619302 0.27571 3270 7613
0.1 53238604 0.390241 1 32462699 53238604 0.41122 4268 12394
0.2 106477208 0.543149 1 48644210 106477208 0.558617 5215 18573
0.3 159715808 0.625887 1 59751787 159715808 0.638094 5693 22814
0.4 212954416 0.67839 1 68488171 212954416 0.688522 5995 26150
0.5 266193008 0.714813 1 75914701 266193008 0.723539 6204 28985
0.6 319431616 0.741736 1 82497808 319431616 0.749427 6378 31499
0.7 372670208 0.76249 1 88513055 372670208 0.769402 6517 33795
0.8 425908832 0.779116 1 94076279 425908832 0.78542 6642 35920
0.9 479147392 0.792733 1 99311385 479147392 0.798541 6747 37918
1 532386027 0.804159 1 104262941 532386027 0.809561 6840 39472 cat /path/to/output/08.report/SS200000135TL_D1.statistics.json
{
"version": "version_v2",
"1.Filter_and_Map": {
"1.1.Adapter_Filter": [
{
"Sample_id": "E100026571_L01_trim_read_1",
"getCIDPositionMap_uniqCIDTypes": "645784920",
"total_reads": "1002214171",
"mapped_reads": "826344259(82.45%)",
"CID_misOverlap_reads": "143590127(14.33%)",