To address these challenges, Stereo-seq OMNI Transcriptomics for FFPE Solution V1.1 was developed as an upgraded spatial transcriptomics solution for FFPE samples. It improves sensitivity, spatial accuracy, and data efficiency, making FFPE-based spatial studies more reliable and accessible for the broader scientific community.
Why FFPE Samples Are Challenging
FFPE samples are valuable because they enable retrospective studies and large clinical cohorts. However, traditional spatial transcriptomics methods often struggle with:
Low RNA capture efficiency
Loss of spatial accuracy due to RNA diffusion
High sequencing waste and cost
Limited flexibility when handling different sample types
Stereo-seq OMNI V1.1 is designed to overcome these limitations.
Key Improvements in Stereo-seq OMNI V1.1
Deeper Insights Per Run - Higher Gene Capture Efficiency
Stereo-seq OMNI V1.1 captures both coding and non-coding RNA, as well as microbial transcripts. With gene capture efficiency shows a 1X–3X increase across tested FFPE tissues, allows researchers to uncover richer RNA profiling and deeper biological insights from every run.
True Single-Cell Profiling at Scale with Preserved Spatial Fidelity
RNA diffusion is a major challenge in FFPE samples. Stereo-seq OMNI V1.1 addresses this with enhanced tissue diffusion control, which retains preserved spatial fidelity at the single-cell level. By minimizing signal spread, it delivers precise true single-cell resolution. This enables clear interpretation of tissue structure and cell organization, allowing for well-defined tissue annotation and stratification (Figure 1).
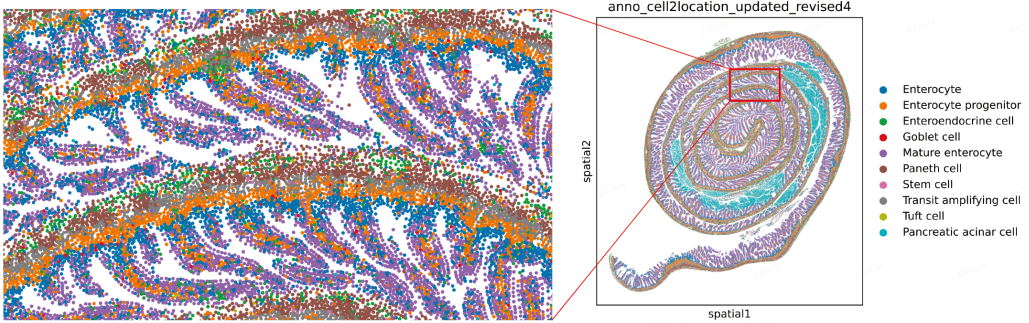
Figure 1. This image displays the improved spatial localization at the nanoscale for key intestinal cell types — including stem cells, transit amplifying cells, enterocyte progenitors, enterocytes, mature enterocytes, goblet cells, Paneth cells, enteroendocrine cells, tuft cells, and pancreatic acinar cells — within the villus crypt architecture, as achieved by the Stereo-seq OMNI V1.1 method
Improved Data Usability - Higher Valid Data Yield
With an improved library construction process and updated bioinformatics tools, Stereo-seq OMNI V1.1 significantly increases the proportion of valid data. Internal evaluations on representative FFPE clinical samples showed an improvement in data usability of over 100%. More raw sequencing reads are retained as valid data, enabling more reliable downstream biological interpretation.
Greater Experimental Flexibility
Stereo-seq OMNI V1.1 supports both fresh-frozen (FF) and FFPE samples in the same sequencing run. Different sample types can be processed together on a single sequencing chip. This reduces logistical constraints and allows for more versatile experimental designs.
Typical Research Applications
Stereo-seq OMNI V1.1 has been validated across many common clinical and research samples. Typical use cases include:
Cancer research: spatial analysis of lung, breast, liver, and other tumors
Infectious disease studies: host–microbe spatial interactions
Neuroscience: high-resolution spatial profiling of brain tissue
Large cohort studies: tissue microarrays (TMAs) and multi-sample stitching on a single chip
In partner evaluations, Stereo-seq OMNI V1.1 showed approximately 2× higher cDNA yield in several FFPE tissues. Valid CID increased by ~40%, and the number of detected genes per bin also doubled, especially benefiting small biopsy samples.
3 Essentials for FFPE Sample Preparation Success
High-quality data starts with high-quality samples. While Stereo-seq OMNI V1.1 is designed to be robust, following these core principles will help ensure the best results:
Block Integrity: Ensure FFPE blocks are well-embedded, free of cracks, and stored properly. Tissue size should fit within the recommended chip area.
Section Quality: Use flat, intact sections (typically 5 μm) with minimal folding or tearing
Careful Handling: Maintain 2–8°C during storage and transport, and clearly distinguish the A/B sides of the chip to prevent tissue detachment during processing
Need the full protocol? Download the Complete FFPE Sample Preparation Guide
Looking Ahead with Stereo-seq OMNI V1.1
Stereo-seq OMNI V1.1 is an important advance for spatial transcriptomics using FFPE samples. It improves gene capture efficiency, spatial accuracy, data usability, and compatibility with archived tissues. Together, these improvements make spatial transcriptomic analysis more reliable and more scalable. For researchers and clincians working with clinical FFPE samples, Stereo-seq OMNI V1.1 enables deeper exploration of complex biological systems that were previously difficult to study.
If you are interested in learning more about Stereo-seq OMNI V1.1, discussing experimental design, or exploring customized spatial transcriptomics solutions, please Contact Us or email us at info_global@stomics.tech.
