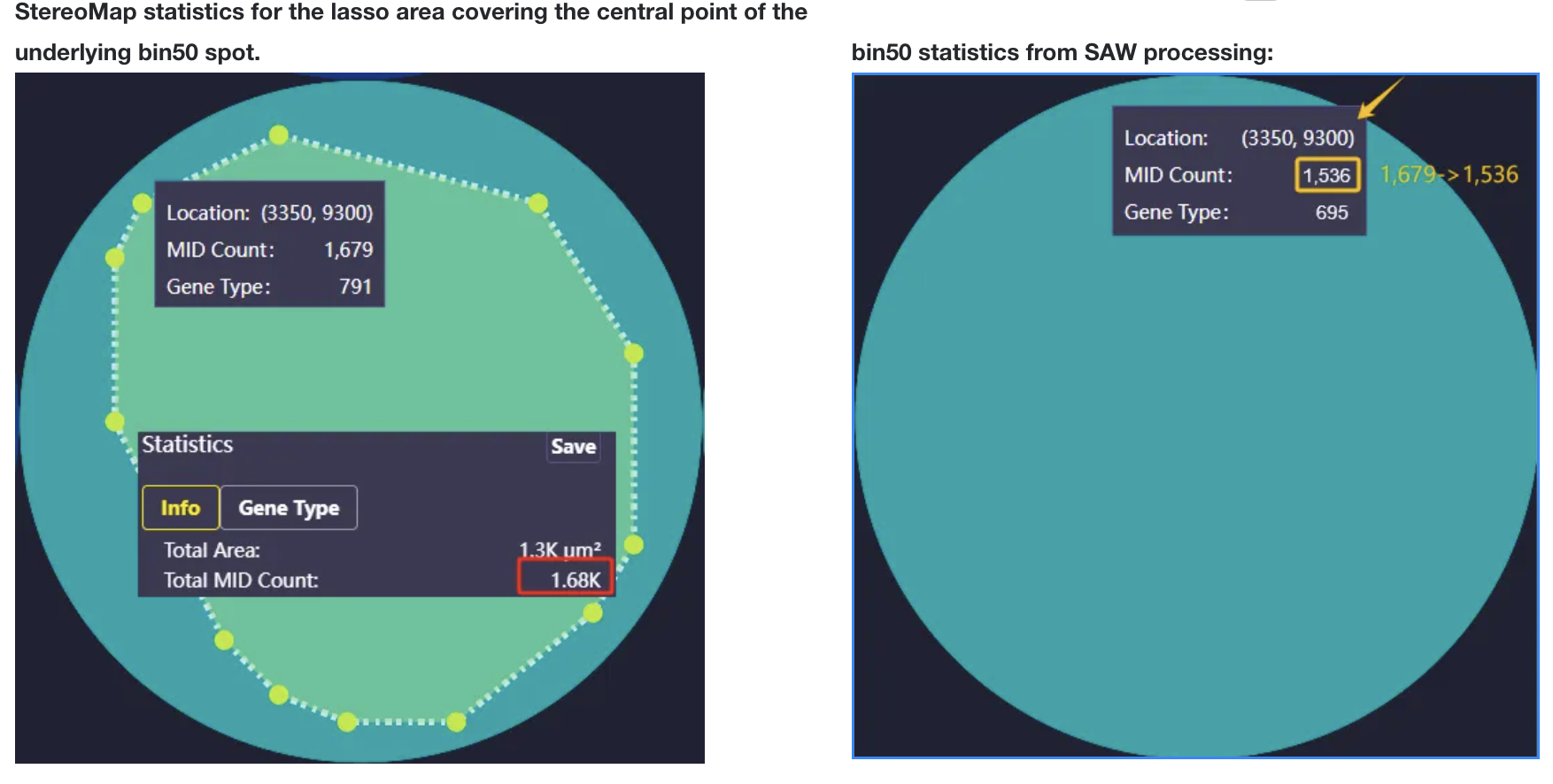
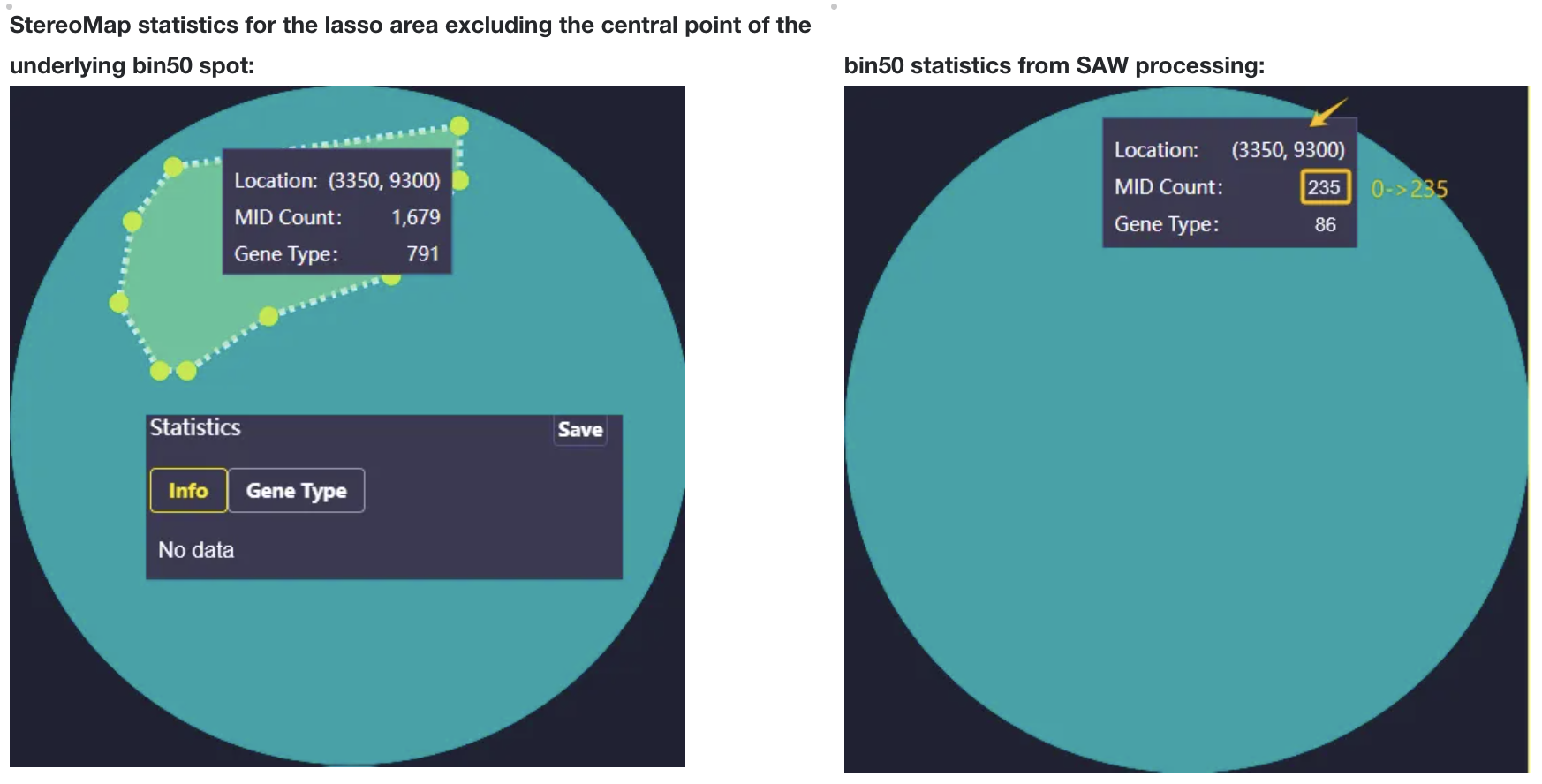
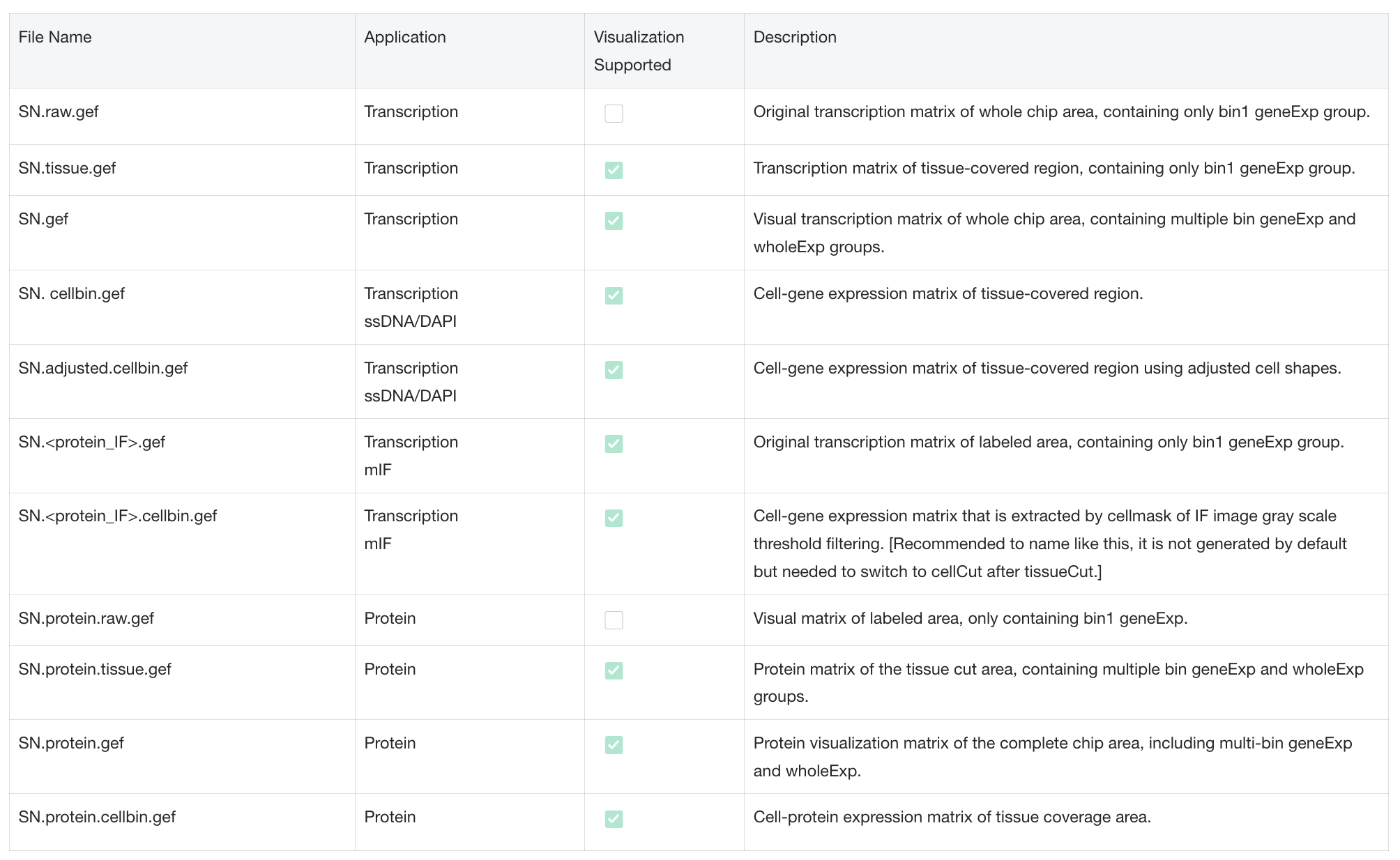
- Bin is a regular NxN square area on the chip. The summary of expression information in the area is the expression information of Bin.
- The values of Bin20, 50, 100 and 200 can be adjusted several times according to the cell size of different tissue types and the downstream analysis effect. Where Bin20 is similar to the size of animal cells, Bin50 and Bin100 are Binsize sizes commonly used for analysis. Bin200 is generally used for quick visualization.
All
Products
Resources
News
FAQ
Search