All
Products
Resources
News
FAQ
Search
09/09/2025 Yahui Li
Neuroendocrine tumors (NETs) originate from specialized secretory cells of the neuroendocrine system and present diverse clinical manifestations. Among these, colorectal NETs (CRNETs) are among the most common subtypes and frequently develop distant metastases, most often in the liver. Over half of CRNET patients exhibit liver metastases (LM) at initial diagnosis, and incidence increases over time. Patients with LM (CRNET-LM) face a significantly worse prognosis, with a 5-year survival rate of only 13–54% compared to 75–80% for localized disease [1]. These findingsunderscore the urgent need to improve the understanding and treatment of CRNET-LM.
Therefore, a recent study published in Molecular Cancer [1] employs single-cell technologies to investigate tumor microenvironment (TME) heterogeneity between primary tumors and liver metastases in CRNET-LM and constructs a single-cell atlas for CRNET tumor samples. The goal is to provide new insights and research avenues for enhancing the prognosis of CRNET-LM.
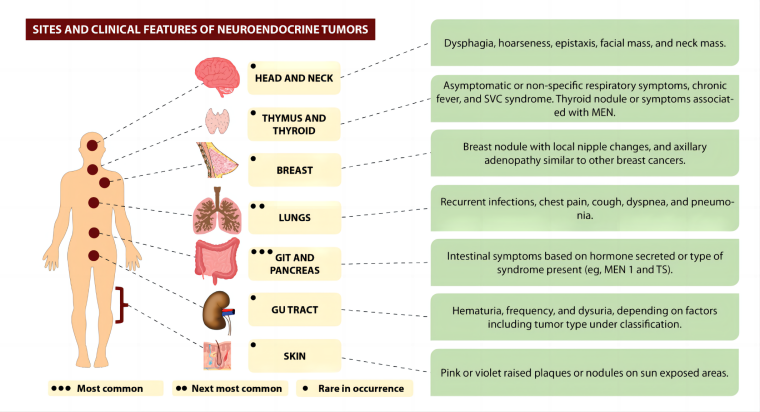
Figure 1 from Sultana et al.[2] illustrates the sites and clinical features of neuroendocrine tumors (NETs), where colorectum is one of the most common sites.
The overall experimental design of this study is as follows: Samples were collected from primary lesions (PL, n=5) and liver metastases (LM, n=4) of 9 CRNET-LM patients. First, comprehensive single-cell transcriptomic analysis was performed to characterize the single-cell atlas of both sites. This was followed by integrated data validation from spatial transcriptomics analysis, multiplex immunohistochemistry, and bulk RNA-seq cohort data from the public GEO database. The aim was to identify cellular and molecular signatures associated with CRNET liver metastasis and potential targets linked to patient survival.
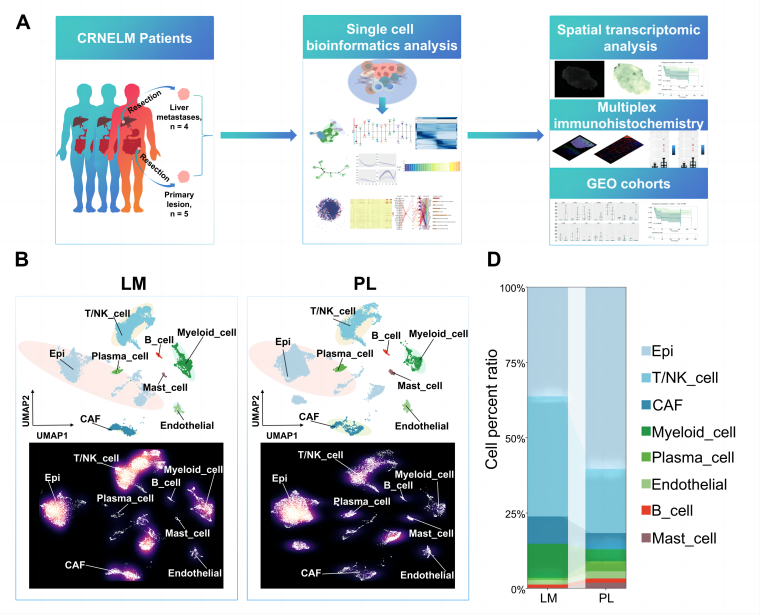
Figure 1 from Deng et al. [1] shows the experimental design and the single-cell landscape of PL and LM of CRNE-LM patients.
Through comprehensive single-cell analysis, including cell type annotation, differential cell proportion analysis, functional and pathway enrichment, trajectory inference, and cell communication analysis, the following findings were obtained:
First, a single-cell atlas was constructed for both LM and PL sites in CRNET, identifying eight major cell clusters, including immune cells (T/NK cells, B cells), cancer-associated fibroblasts (CAFs), and myeloid cells. Further analysis revealed differences in cell cluster proportions between the two sites: epithelial cells, plasma cells, endothelial cells, and mast cells were more abundant in PL, whereas T/NK cells, myeloid cells, CAFs, and B cells were proportionally higher in LM.
Secondly, subtype analysis of individual cell categories revealed that within the tumor microenvironment (TME) of LM sites, CD8⁺ T cells, NK/NKT cells, CD4⁺ T cells, and B cells all exhibited a more pronounced stress-like phenotype. As the stress score increased, these immune cells showed a significant rise in inhibitory scores, accompanied by a decline in cytotoxicity scores in CD8⁺ T, NK, and NKT cells. These findings suggest a potential mechanism of immune resistance in metastatic neuroendocrine tumors. In contrast, such features were not observed in immune cells from PL sites. The prevalent stress-like phenotype in the liver metastatic TME may be a key factor contributing to the poor response of CRNET-LM to immunotherapy.
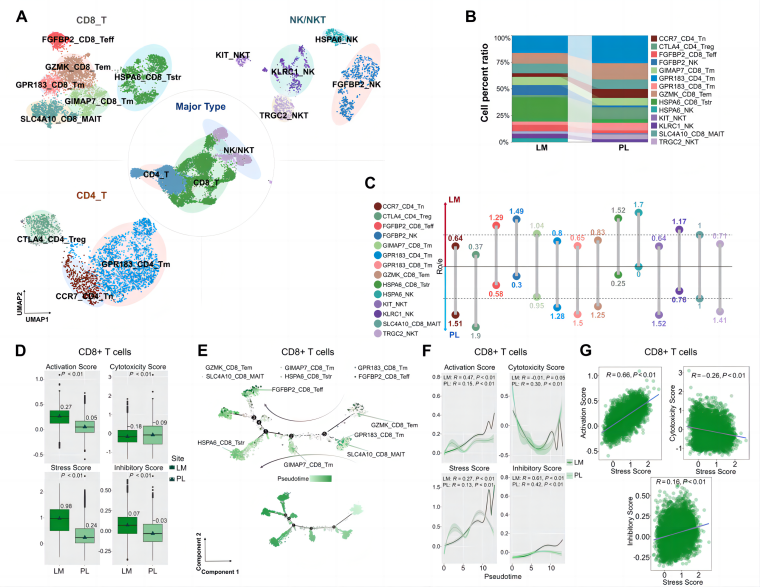
Figure 2A-G from Deng et al [1] demonstrates the subtypes of T, NK, and NKT cells between PL and LM of CRNELM patients.
The study also revealed that macrophages in primary tumors (PL_MCs) and those in liver metastases (LM_MCs) exhibit distinct activation patterns of tumor-associated signaling pathways. Notably, COLEC11⁺ matrix cancer-associated fibroblasts (COLEC11+ mCAFs) showed a significant correlation with LM_MCs, but no significant positive correlation was observed with PL_MCs in primary lesions. Cell communication analysis identified unique receptor–ligand interactions and activated pathways between COLEC11+ mCAFs and LM_MCs, which differed from those involving PL_MCs. Targeting these specific receptors or ligands in liver metastases may offer a potential therapeutic strategy for modulating cellular crosstalk and immune responses.
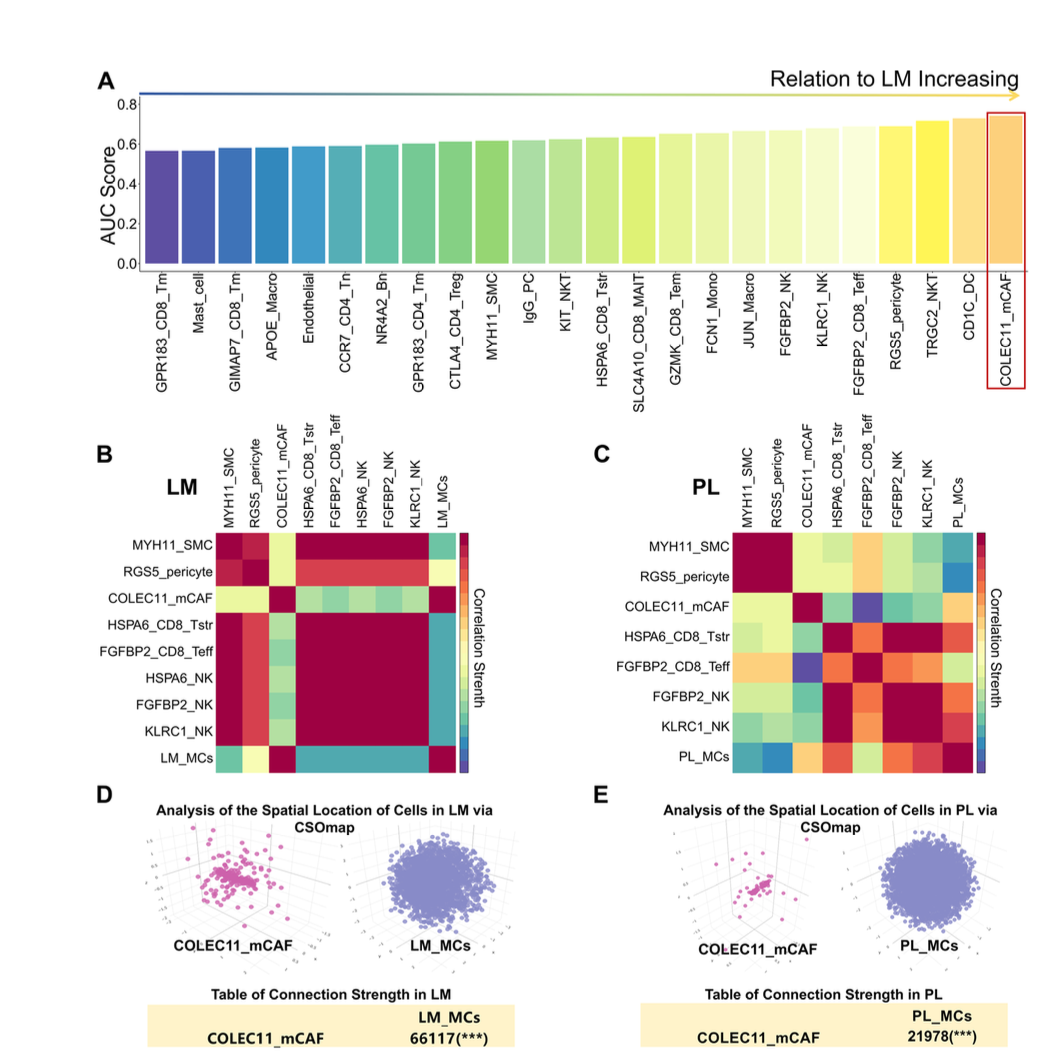
Figure 7A-E from Deng et al. [1] shows the pivotal role of COLEC11_mCAFs in supporting liver metastasis-associated macrophages (LM_MCs) in the TME at the metastasis site.
To further validate the spatial distribution differences of COLEC11_mCAFs, the team applied Stereo-seq, a high-resolution sequencing-based spatial transcriptomics technology, to analyze paired primary (PL) and liver metastasis (LM) samples, precisely mapping the spatial distribution patterns of different cell subpopulations within the TME. The analysis revealed a higher abundance of COLEC11_mCAFs in LM compared to PL. This finding was further validated through gene set-based scoring, bulk RNA-seq data from the GEO database, and multiplex immunohistochemistry/immunofluorescence (mIHC/IF) experiments. Moreover, patient cohorts with high COLEC11_mCAF abundance exhibited poorer overall survival, suggesting that targeting this specific cancer-associated fibroblast subpopulation may represent a potential therapeutic strategy for CRNET-LM patients.
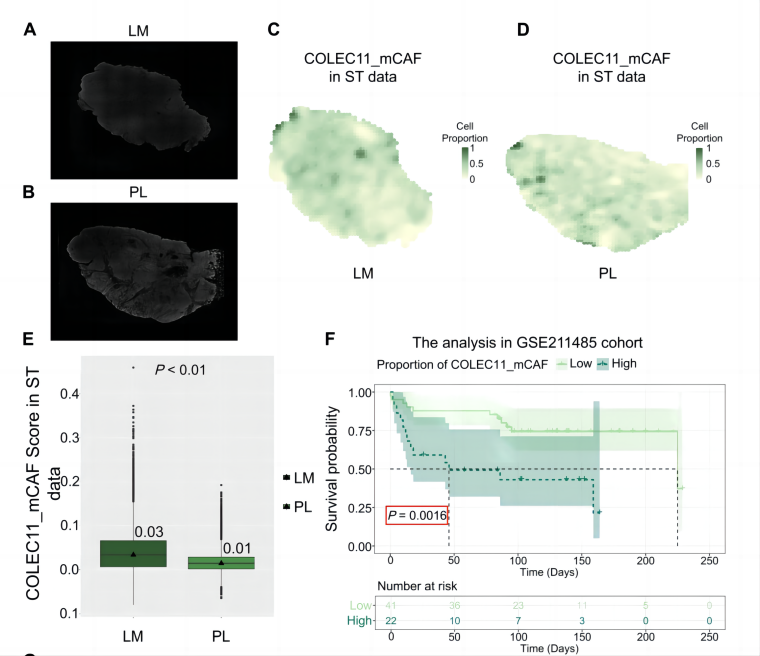
Figure 8A-E. from Deng et al. [1] demonstrates increased enrichment of COLEC11_mCAFs in liver metastases (LM) compared to primary lesions (PL), as revealed by spatial transcriptomics.
In summary, this study provides the first single-cell perspective on the cellular and molecular heterogeneity between primary tumors and liver metastases in CRNET-LM patients. It identifies specific cell subpopulations and associated receptor–ligand interactions that may help explain the distinct features of the CRNET-LM tumor microenvironment, offering a basis for potential therapeutic targets and improved management strategies. Notably, the application of Stereo-seq spatial transcriptomics enabled high-resolution, large-field spatial mapping of cell subtypes, which further validated the single-cell findings and underscored the value of this technology in biomedical research.
1. Deng, Y., et al. Comprehensive single-cell atlas of colorectal neuroendocrine tumors with liver metastases: unraveling tumor microenvironment heterogeneity between primary lesions and metastases. Mol Cancer (2025). https://doi.org/10.1186/s12943-025-02231-y
2. Sultana Q., et al. A Comprehensive Review on Neuroendocrine Neoplasms: Presentation, Pathophysiology and Management. J Clin Med. (2023).
https://pmc.ncbi.nlm.nih.gov/articles/PMC10420169/
Author:
![]() Dr. Yahui Li, Field Application Scientist (Bioinformatics) at STOmics Tech
Dr. Yahui Li, Field Application Scientist (Bioinformatics) at STOmics Tech