All
Products
Resources
News
FAQ
Search
30/09/2025 Can Shi, Mei Li, Ying Zhang
Note: obtain and use CSRefiner: https://github.com/STOmics/CSRefiner
Cell segmentation accuracy is critical for reliable single-cell spatial transcriptome data, directly impacting subsequent gene expression and spatial analysis credibility. For Stereo-seq data, we first recommend CellBin (see Blog [1]), a SAW-embedded tool and a deep-learning model trained/optimized on large Stereo-seq datasets, offering targeted, high-accuracy, large-field-of-view segmentation. We also evaluated other mainstream models on stained multi-tissue images, providing recommendations based on accuracy/robustness to match user-specific image features (see Blog [2]).
However, end-users prioritize "accurate segmentation of their own data" over statistical overall accuracy. Even with >95% global accuracy, local under-segmentation (missed cells) or over-segmentation (background misidentified as cells or divided a cell into multiple cells) causes critical errors. These undermine differential expression analysis, rare cell subpopulation identification, and cell spatial interaction network construction—even leading to incorrect biological conclusions.
Manual annotation is the ultimate cell segmentation solution but labor-intensive: a 1 cm × 1 cm sample may take an annotator >2 weeks, increasing costs and delaying research. Thus, we suggest combining local manual annotation with automatic fine-tuning via CSRefiner—a lightweight framework for fine-tuning cell segmentation models with small datasets for spatial transcriptome pipelines.
Ideal for large-batch sample projects, CSRefiner currently supports three leading models in spatial omics: Cellpose, StarDist, and CellBin (more models planned). With minimal annotated local data, it fine-tunes existing models to specific image features; the fine-tuned model then segments entire/similar images, boosting efficiency while ensuring key-region accuracy.
As illustrated in Figure 1, the workflow of CSRefiner comprises the following key steps:
Users construct a small-scale training set by annotating representative patches in regions where the baseline model exhibits poor segmentation performance (e.g., dense regions or regions with morphological heterogeneity, such as the hippocampus). Subsequently, a base model is selected from the supported models (Cellpose, StarDist, or CellBin).
The chosen model is fine-tuned on the annotated training set using a unified, parameter-controlled script, yielding a new fine-tuned model adapted to the features of the target image.
The refined model is evaluated on the training set and testing set. If necessary, the model is iteratively refined by adding more annotated patches—this ensures the model achieves satisfactory performance across challenging regions.
The optimized model is applied to whole-slide images. The resulting segmentation boundaries are integrated with the corresponding expression matrix to generate a single-cell-level gene expression format (cgef) file. As a widely used standard in spatial transcriptomics, this file facilitates downstream single-cell analyses.
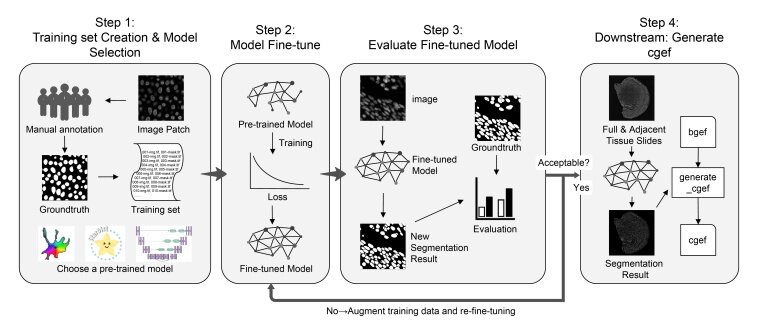
Figure 1. Schematic overview of the CSRefiner workflow
To intuitively demonstrate the improvement of CSRefiner in practical applications, we selected a representative Stereo-seq FFPE DAPI-stained mouse brain dataset. We fine-tuned the four models (Cellpose-cyto, Cellpose-cpsam, StarDist, CellBin) on this dataset and compared its cell segmentation performance before and after fine-tuning.
As shown in Figure 2A, we observed significant inconsistencies in model performance across different tissue regions: in non-hippocampal regions with sparse cells and clear nuclear boundaries, the model typically captured cell contours. However, in the hippocampus (characterized by dense cells and blurred nuclear boundaries), all pre-trained models exhibited obvious defects.
As illustrated in Figure 2B, after fine-tuning with CSRefiner, in non-hippocampal regions, more precise segmentation was achieved, closely matching the actual nuclear contours with few missed cells. In the hippocampus, pre-trained models suffered from severe false negatives due to dense cell clustering and blurred nuclear boundaries, while CSRefiner showed remarkable improvement: it successfully recovered many previously missed cells and could distinguish individual nuclei even in the densest cell clusters.
 Representative segmentation results from four pre-trained models (Cellpose-cyto, Cellpose.jpg)
Figure 2. (A) Representative segmentation results from four pre-trained models (Cellpose-cyto, Cellpose-cpsam, StarDist, and CellBin). Red contours indicate ground-truth manual annotations, and yellow contours indicate model-predicted segmentation boundaries. (B) Segmentation results after fine-tuning with CSRefiner, corresponding to the regions shown in panel A. Model names prefixed with “FT-” indicate fine-tuned versions (same hereafter).
As presented in Figures 3A–E, we evaluated CSRefiner's impact on segmentation accuracy using five standard evaluation metrics: Precision, Recall, F1-score, Jaccard Index, and Dice Coefficient. Box plot comparisons revealed that all metrics of all tested models were improved. Notably, this improvement was particularly significant for models with initially weak performance, while even the high-performance Cellpose-cpsam model achieved measurable gains. Additionally, the reduced variance in scores after fine-tuning indicated enhanced consistency and robustness of the model across different image regions.
 Quantitative evaluation of segmentation performance before and after fine-tuning across.jpg)
Figure 3. (A–E) Quantitative evaluation of segmentation performance before and after fine-tuning across four representative models. Boxplots show improvements in (A) precision, (B) recall, (C) F1 score, (D) Jaccard index, and (E) Dice coefficient. The significance mark line and P value are added to the box plot. (F) Time required for manual whole-slide annotation (~10 days) versus CSRefiner-assisted workflow (~400 minutes).
As shown in Figure 3F, CSRefiner drastically shortened annotation time. Based on an average annotation time of ~15 minutes per 256 × 256 image patch, generating 20 training patches required ~300 minutes. Model fine-tuning, evaluation, and inference added an additional ~100 minutes, resulting in a total of ~400 minutes (~6.7 hours) for a complete CSRefiner workflow. In contrast, manual annotation of an entire whole-slide image would take approximately 10 days (~36× longer) under comparable conditions.
To assess CSRefiner’s biological relevance, we analyzed how its enhanced segmentation accuracy affects downstream spatial transcriptomics analyses. Using StarDist (the model with the largest performance gain), fine-tuned segmentation generated high-quality cgef matrices; subsequent cell type annotation via cell2location produced spatial distributions that closely matched established anatomical structures and biological priors (Fig. 4A). Notably, hippocampal subregions were delineated with high fidelity (Fig. 4B), confirming that CSRefiner-generated segmentations directly support biologically meaningful spatial mapping in downstream analyses.
 Spatial maps of cell type annotations generated by cell2location using cgef matrices from.jpg)
Figure 4. (A) Spatial maps of cell type annotations generated by cell2location using cgef matrices from the fine-tuned StarDist model. (B) Visualization of segmented and annotated cells in hippocampal subregions before and after fine-tuning compared with the Allan Brain Atlas for the hippocampus.
CSRefiner is a promising tool for spatial transcriptomics studies due to its efficiency and flexibility in obtaining more accurate cell segmentation results. We encourage the community to adopt CSRefiner for their research and refer readers to our preprint [3] for detailed results and technical insights.
[1] CellBin: The Core Image Processing Pipeline in SAW for Generating Single-cell Gene Expression Data for Stereo-seq https://en.stomics.tech/news/stomics-blog/1017.html
[2] Community Tool Recommendations for Cell Segmentation in Stereo-seq Data https://en.stomics.tech/news/stomics-blog/1162.html
[3] Shi, Can, et al. "CSRefiner: A lightweight framework for fine-tuning cell segmentation models with small datasets." bioRxiv (2025): 2025-09.