All
Products
Resources
News
FAQ
Search
24/01/2025 Yahui Li, Ying Zhang, Mei Li
Stereo-seq is a sequencing-based spatial omics technology developed by STOmics. Since its first publication in 2022 [1], it has been successfully applied across a range of research fields, including organ development, oncology, tissue regeneration, and disease research. Stereo-seq enables single-cell resolution spatial transcriptomics analysis using a DNA nanoball-patterned array. This innovative approach features RNA-capturing spots with a diameter of 220 nm, spaced 500 nm apart on a chip. While this design theoretically allows for spatial analysis at a resolution finer than individual cells, true single-cell resolution is primarily achieved through advanced image processing. CellBin, an image processing pipeline embedded in SAW (Stereo-seq Analysis Workflow), the primary bioinformatics software for Stereo-seq data analysis, plays a crucial role. The pipeline includes several image processing algorithms tailored for various Stereo-seq applications, such as a unique nuclei segmentation algorithm that adapts to different sample staining methods. In this blog, you will gain a fundamental understanding of the CellBin pipeline, its advantages in single-cell resolution spatial transcriptomics analysis, and the diverse scenarios in which it can be applied.
CellBin is an image processing pipeline designed to delineate cell boundaries for spatial analysis. It consists of several image analysis steps. Given the image and gene expression data as input, CellBin performs image registration, tissue segmentation, nuclei segmentation, and molecular labeling (i.e., cell border expanding), ultimately defining the molecular boundaries of individual cells. It incorporates a suite of self-developed algorithms, including deep-learning models, for each of the analysis task. The processed data is then mapped onto the chip to extract molecular information, resulting in an accurate single-cell expression matrix. (Figure 1) For more information on CellBin, please refer to this preprint [2].
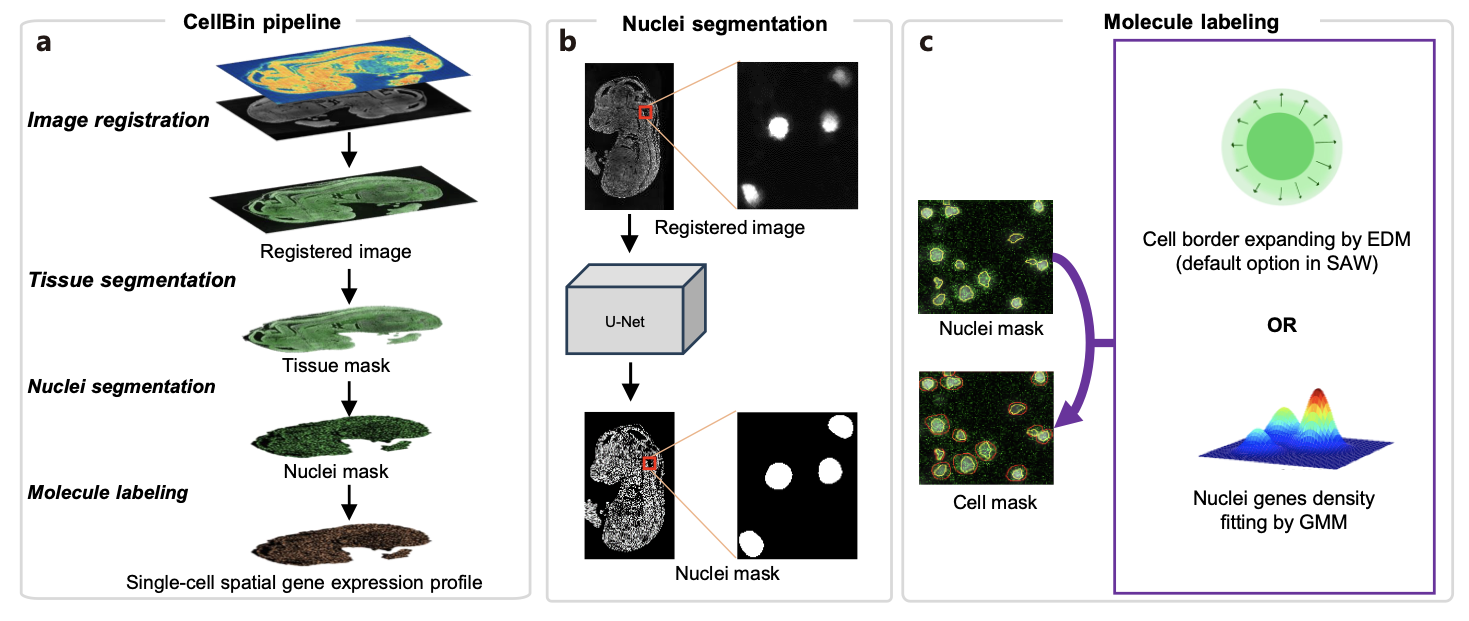
Figure 1. CellBin pipeline and its core algorithms. a. Overview of CellBin pipeline.b. The self-developed nuclei segmentation algorithm. c. The molecule labeling step for more accurate identification of cell borders. EDM: Euclidean Distance Map; GMM: Gaussian Mixture Model. (Figure adapted from [2])
The CellBin pipeline has been wrapped within the SAW software [3], becoming part of the automated workflow (Figure 2). Briefly, sequencing reads, spatial barcode data (i.e., mask), and the reference genome are first pre-processed. The reads are then mapped and annotated to generate a coordinated gene expression matrix. Then CellBin aligns the microscopic images with the gene expression matrix using tracklines on the Stereo-seq chip. Currently, registration error is within 5 μm, which is smaller than the size of a single cell. Next the tissue segmentation and cell segmentation steps are performed to delineate the tissue and cell regions of the tissue based on the images. As a result, SAW can extract gene expression matrix based on both the tissue and the cell regions, enabling downstream bioinformatics analysis.
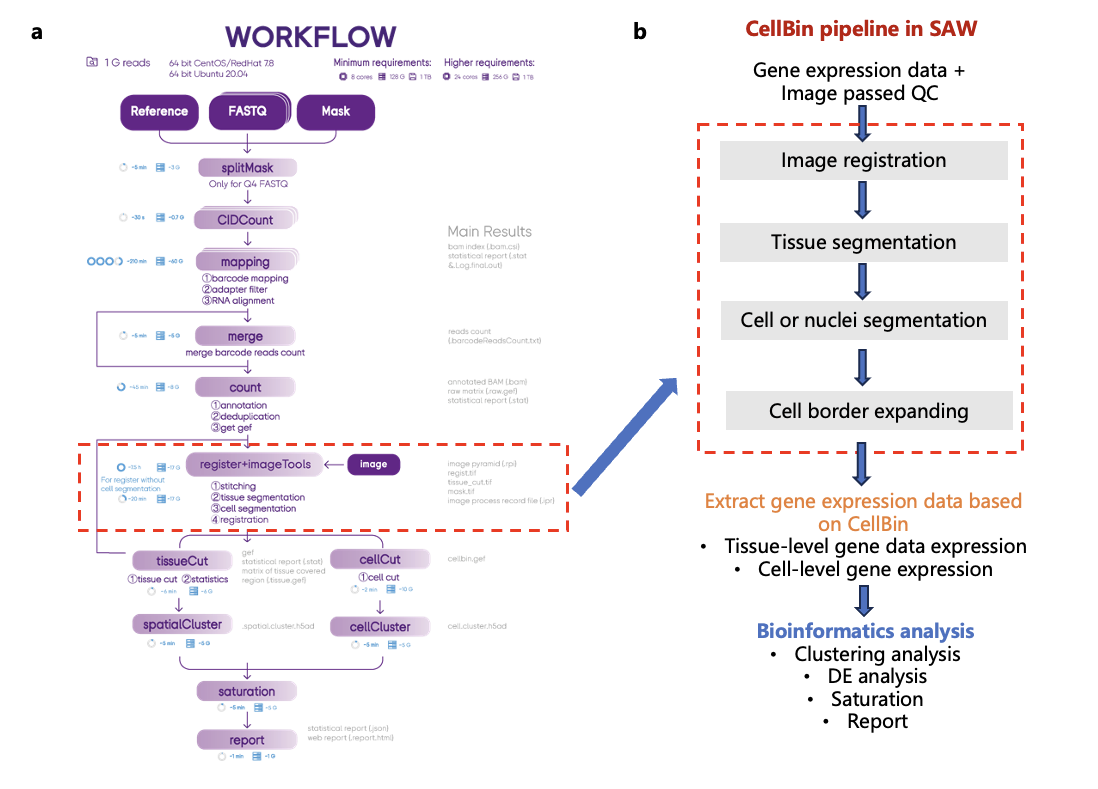
Figure 2. Diagram illustrating the SAW workflow and highlighting the CellBin steps. a. Overview of the SAW workflow. b. CellBin pipeline (red dashed box) within SAW and its downstream steps.
Accurate determination of cell localization and the generation of single-cell expression matrix can significantly enhance the analytical potential of Stereo-seq data. This enables deeper feature analysis and more precise cell type identification, which are crucial for interpreting cell-cell interactions and uncovering underlying biological patterns. Stereo-seq offers two types of analysis units available for downstream analysis, square bin (or bin), and cell bin (or cellbin) (Figure 3). A square bin is based on the chip coordinates and offers a range of size options, such as bin1, bin5, bin20, bin100. A cell bin, on the other hand, represents single-cell unit generated by the CellBin pipeline and requires a microscopic image for nuclei or cell boundary identification. Here, bin20 is a 10μm diameter square that approximates the size of an animal cell. Therefore, bin20 is chosen for performance comparison with the cell bin.
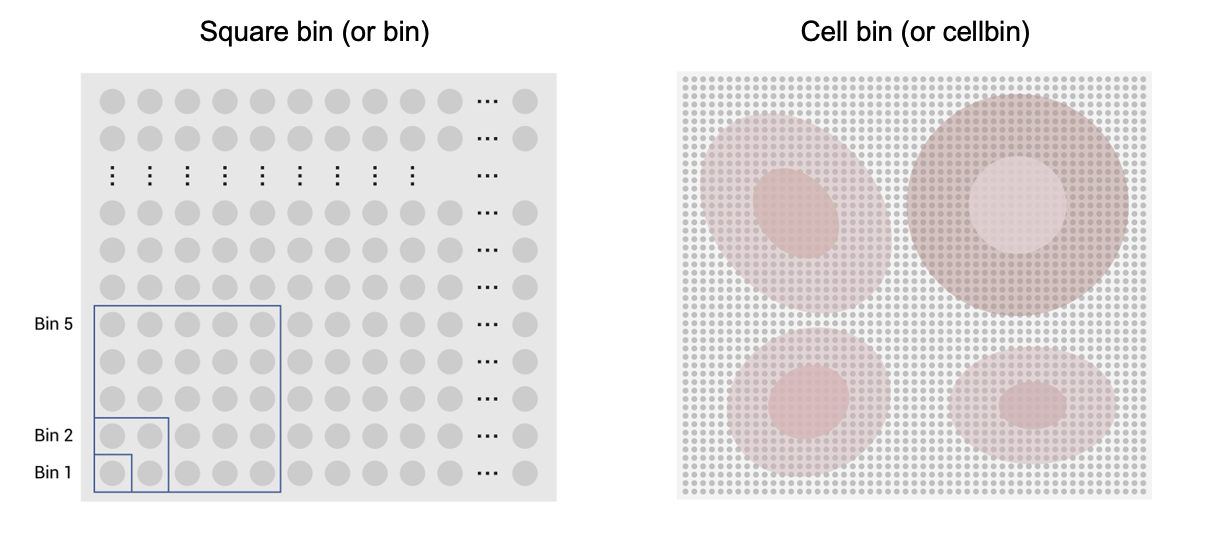
Figure 3. Diagrams of square bin and cell bin. The two types of analysis units are generated by dividing spatial coordinates (square bin) and using the CellBin algorithm (cell bin), respectively.
Using a mouse brain spatial transcriptomics analysis as an example, both CellBin and the bin20 strategies are applied to obtain single-cell expression matrices, and the data analysis results are compared. As expected, CellBin significantly improves cell integrity by minimizing cross-contamination of expression information and preventing the generation of pseudo-cell results (Figure 4).
Downstream analysis reveals several key advantages of using CellBin-derived expression matrix:
i) Higher gene counts. The numbers of molecular identifiers (MIDs) and genes per cellbin are higher than that of bin20 (Figure 5a).
ii) Better clustering results. Cellbin data exhibit a higher number of clusters compared to bin20, and also a lower Davies-Bouldin (DB) score suggesting the clusters are more compact and well-separated. When zooming in the distinct cortical regions, a higher Moran's I index (a score measuring spatial autocorrelation) for cellbin is observed, depicting better clustering result in these regions (Figure 5b-e).
iii) More accurate cell annotation results. Using an end-to-end cell type annotation method, Spatial-ID [4], CellBin data demonstrate more accurate results, with more significant expression of marker genes in the annotated cells (Figure 6).
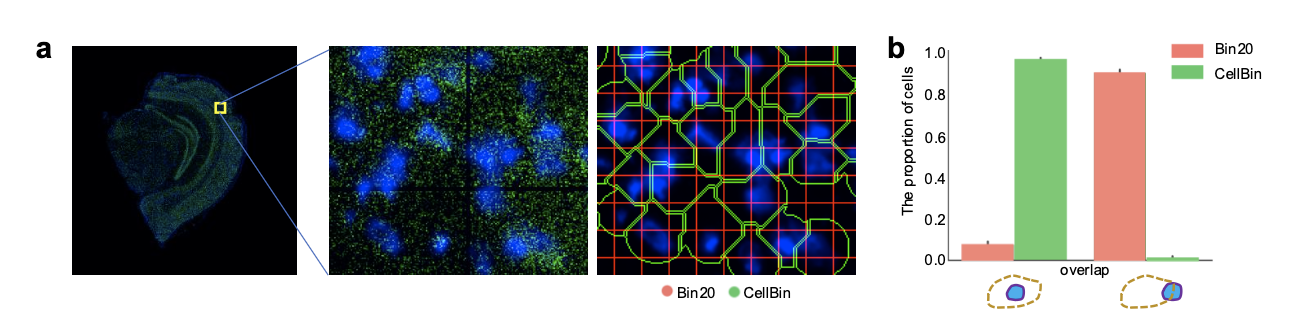
Figure 4. Two strategies for obtaining single-cell expression matrices. a. Expression matrix overlaid with staining images, showing area divisions by bin20 and CellBin. b. Comparison of cell integrity between the two strategies. (Figure adapted from [2])
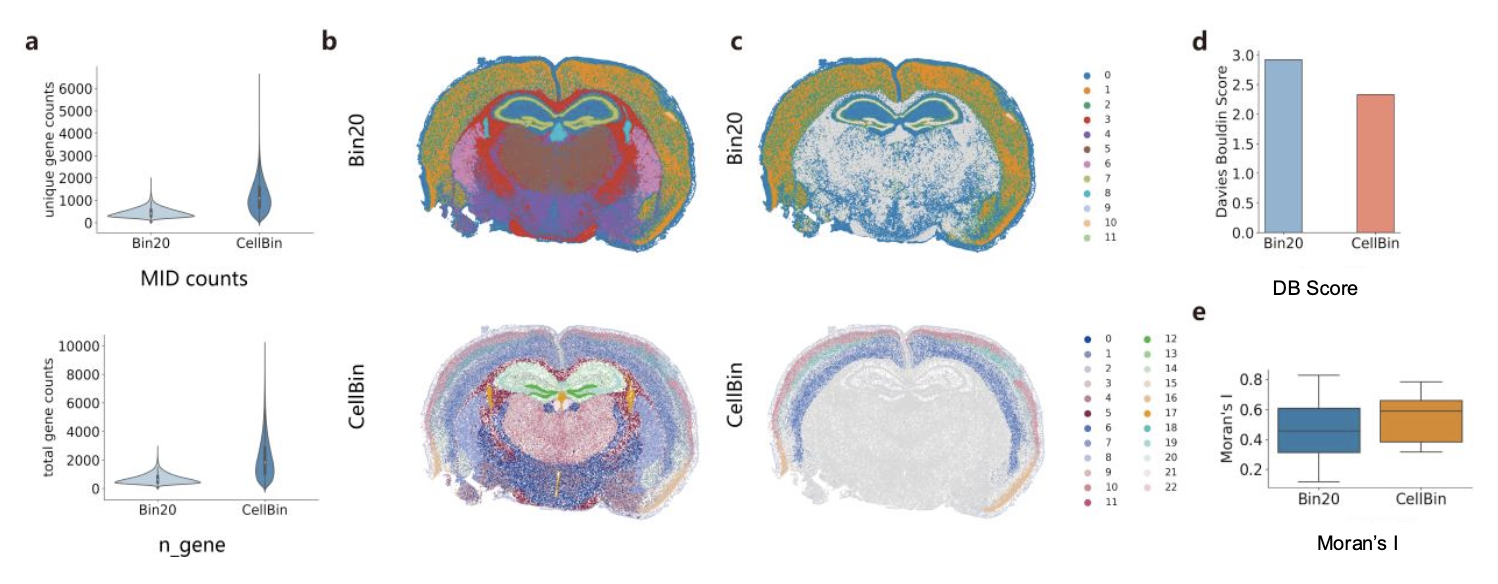
Figure 5. Comparison of downstream analyses between bin20 and CellBin a. Comparison of MID counts and unique gene counts. b. Clustering analysis results using the Leiden algorithm. c. Highlighted clusters corresponding to cortical regions. d. Comparison of the Davies-Bouldin (DB) scores for the clustering results across the whole brain tissue. e. Comparison of Moran's I index for the clustering results in the cortical regions.
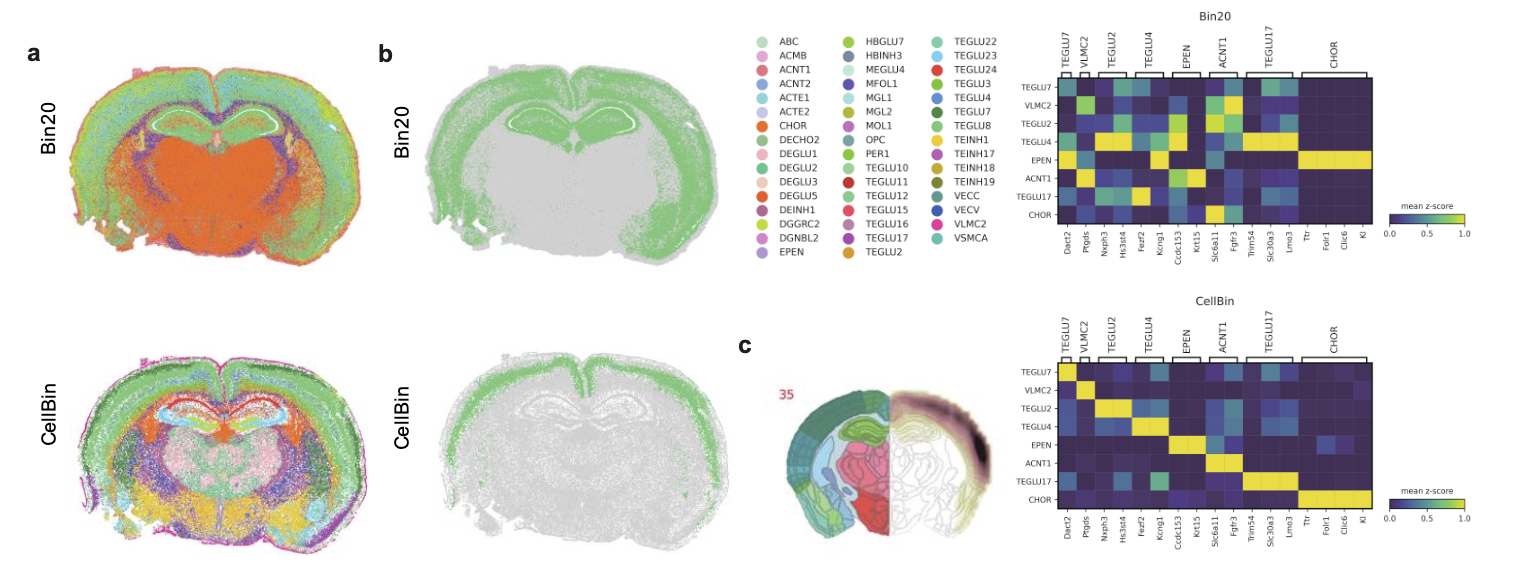
Figure 6. Comparison of cell annotation results between bin20 and CellBin. a. Spatial-ID annotation results. b. Comparison of TEGLU8 cell annotation results. c. Probability distribution of potential locations for TEGLU8 based on Allen Brain Atlas. d. Heatmap showing the expression correlation of marker genes for selected cell types.
The CellBin pipeline is designed to be compatible with versatile Stereo-seq products, and therefore can accommodate multiple staining methods, including ssDNA, DAPI, and H&E. The detailed compatibility of CellBin on Stereo-seq products has been tested by STOmics R&D team and is listed in Table 1. Beyond these mentioned staining methods, CellBin also supports calcofluor white (CFW) staining for plant cell wall staining [5], though this application has not yet been developed into a commercial product. Furthermore, CellBin is under continuous development to expand its compatibility with a wider range of staining methods and tissue types. The developing version can be used as a stand-alone python pipeline. For more details, please refer to the GitHub page for more details [6].
Table 1. Compatibility of CellBin pipeline with Stereo-seq applications.
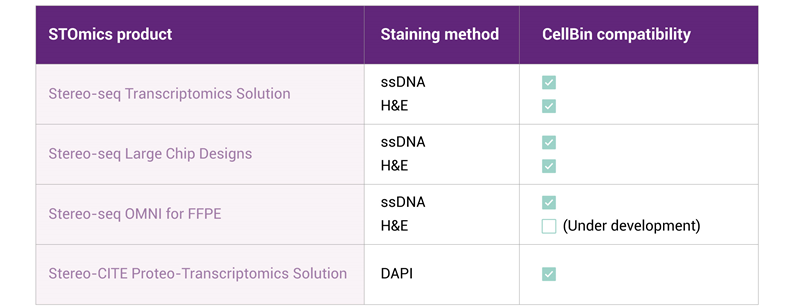
Notes: 1. All current Stereo-seq products, except for the Stereo-seq OMNI solution, are designed for use with fresh frozen (FF) samples; 2. FFPE: Formalin-fixed paraffin-embedded.
CellBin demonstrates high compatibility across tissues from multiple species, including both animal tissues and plant tissues. Figure 7 showcases CellBin’s performance on a variety of samples, including mouse testis, brain, olfactory bulb, and Arabidopsis seed. In addition, CellBin has been successfully applied to a wide range of tissue types, such as rat brain, zebrafish heart, pig uterus, and human cancer tissues. To explore our publicly available examples, please visit the "Demo data" section on the STOmics website.
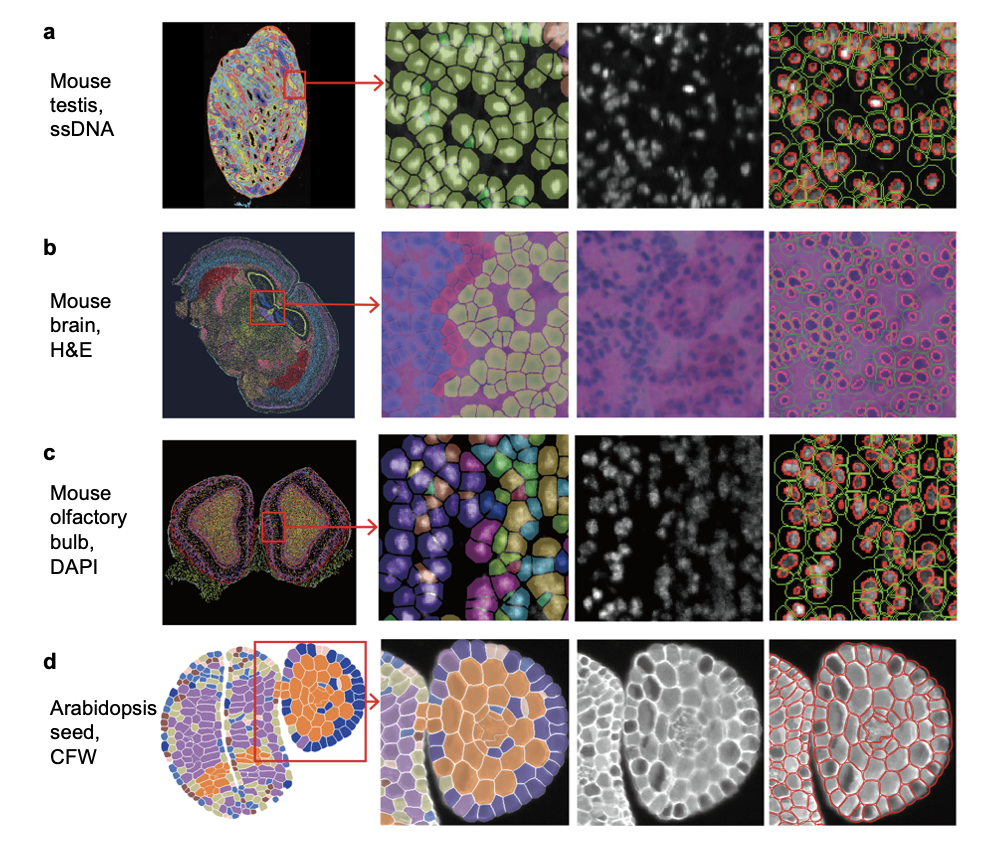
Figure 7. Examples of cell segmentation results using CellBin. Columns from left to right: 1. Cell clusters; 2. Zoomed-in view of clusters; 3. Tissue microscopic image; 4. Cell segmentation result. The red lines denote the nuclei or cells, and the green lines indicate the adjusted cell borders. (Figure 7d adapted from [5])
The CellBin pipeline is a powerful tool for obtaining single-cell spatial gene expression data. However, there are several key factors that must be in place for it to function properly, including image quality, tissue properties, and gene capture efficiency. For high-quality microscopic imaging, it is important not only to ensure that the tissues and cells (or nuclei) are clear, but also to verify the tracklines on the Stereo-seq chip are visible and distinct. These tracklines are crucial for CellBin to align gene expression data with the microscopic images. Since CellBin relies on a deep learning model for nuclei segmentation, it performs best with tissue types that are similar to those it was trained on. For tissue with irregular cell shapes or those not included in the training dataset, performance may be less reliable. Additionally, to generate meaningful results in downstream analyses, it is essential to capture a sufficient number of molecules or genes. Typically, when the median number of genes per bin20 exceeds 200 and image quality is high, analyzing data at the single-cell level (cellbin unit) is recommended. However, when gene capture is suboptimal, larger analysis units, such as bin50 or bin100, should be used instead.
In cases where CellBin cannot be effectively applied or its performance is suboptimal, manual image processing in StereoMap is advised. The image processing module in StereoMap is designed for step-by-step manual image registration and segmentation, and it also supports incorporating segmentation results from third-party tools. After completing the manual processing, the SAW analysis can be re-run. A tutorial on this process is available in the SAW user manual.
In summary, CellBin is the analysis pipeline that allows single-cell spatial analysis for Stereo-seq, and it offers significant improvements in single-cell level gene expression analysis compared to traditional square bin method, ensuring higher cell integrity and yielding better downstream analysis results. CellBin continues to evolve to enhance its compatibility with broader range of Stereo-seq applications and improve its performance for more complex tissue scenarios. Lastly, we would love to hear your thoughts and experiences with CellBin analysis. Feel free to reach out for further discussion or if you have any questions at info_global@stomics.tech.
1. Chen, A., Liao, S., Cheng, M., Ma, K., Wu, L., Lai, Y., et al. (2022). Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell, 185(10), 1777-1792.e1721. doi:10.1016/j.cell.2022.04.003
2. Li, M., Liu, H., Kang, Q., Fang, S., Li, M., Zhang, J., et al. (2024). CellBin: a highly accurate single-cell gene expression processing pipeline for high-resolution spatial transcriptomics. bioRxiv, 2023.2002.2028.530414. doi:10.1101/2023.02.28.530414
3. SAW (V8.1.2 by Jan 23, 2025) user manual gitbook: https://stereotoolss-organization.gitbook.io/saw-user-manual-v8.1
4. Shen, R., Liu, L., Wu, Z., Zhang, Y., Yuan, Z., Guo, J., et al. (2022). Spatial-ID: a cell typing method for spatially resolved transcriptomics via transfer learning and spatial embedding. Nature Communications, 13(1), 7640. doi:10.1038/s41467-022-35288-0
5. Zhang, B., Li, M., Kang, Q., Deng, Z., Qin, H., Su, K., et al. (2024) Generating single-cell gene expression profiles for high-resolution spatial transcriptomics based on cell boundary images. GigaByte. 2024 Feb 20;2024:gigabyte110. doi: 10.46471/gigabyte.110
6. CellBin2 GitHub page: https://github.com/STOmics/cellbin2