All
Products
Resources
News
FAQ
Search
19/05/2025 Yahui Li, Leying Wang
While Stereo-seq analysis with SAW software is designed for efficiency, many users encounter unexpected hurdles during automated image processing--failed Image QC, misaligned registrations, and segmentation errors--which can significantly impact analysis timelines. Building on our previous blog "Stereo-seq Image Processing: Essential Steps and Their Key Points", which detailed core image processing steps, we now explore practical solutions to common analysis challenges using StereoMap's tools and third-party software integration.
When manual adjustments become necessary after careful evaluation on SAW outputs, an additional manual image processing step by StereoMap is integrated into the workflow. This is then followed by the SAW realign pipeline to complete the analysis, as illustrated in Figure 1. The realign pipeline serves the crucial function of continuing the analysis where it left off, incorporating the results from the initial SAW count analysis and the newly corrected image files produced by StereoMap's Image Processing module. Through this process, it ultimately generates spatial gene expression data with accurate tissue and cell context.

Figure 1. The workflow of Stereo-seq data analysis incorporating the manual image adjustment step.
Given that each of the four main image processing steps - from Image QC to final cell segmentation - can encounter specific challenges, we have systematically organized potential issues according to the major steps. Table 1 presents this comprehensive troubleshooting guide, detailing various problem scenarios that may arise during image analysis along with corresponding solutions.
Table 1. Common Stereo-seq image processing issues and solutions.
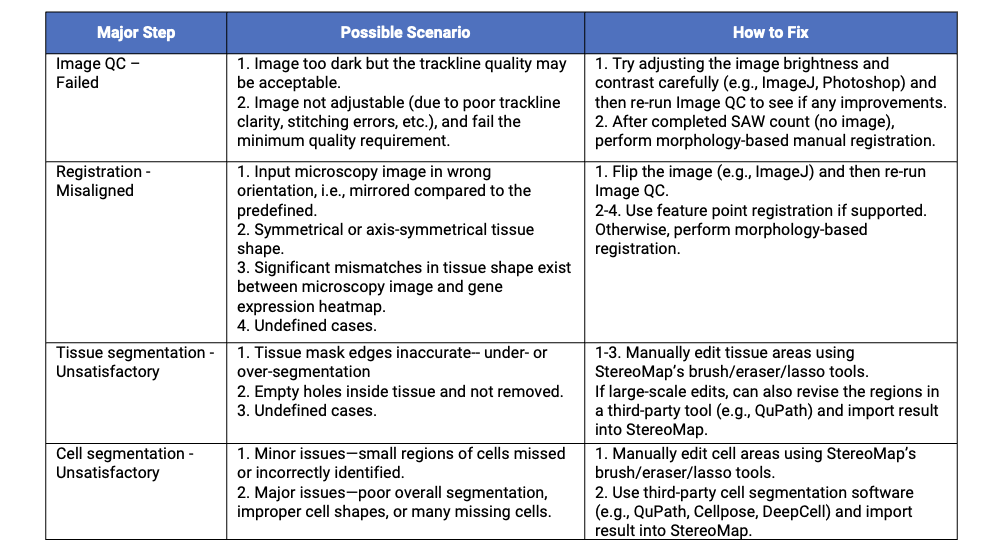
When Image QC fails, this typically indicates insufficient trackline visibility in the microscopy image. If the image is too dark to see the tracklines, users can attempt to enhance trackline detection through careful image adjustments (e.g., brightness/contrast modification) using tools such as ImageJ or Photoshop. However, exercise caution when making these adjustments, as excessive modifications like over-exposure may compromise subsequent tissue and cell boundary identification.
If image is not adjustable and fails the Image QC, run the SAW count pipeline in "no image" mode (omit --image-tar; see the last figure for SAW count command). This skips registration and performs tissue segmentation directly from the gene expression matrix. Next, perform morphology-based manual registration in StereoMap, then export the aligned image for processing in the SAW realign pipeline (automates tissue and cell segmentation). If the automatic segmentation results are sub-optimal, iteratively refine through manual adjustments and SAW realign until all images satisfying.
Unsatisfactory registration occurs when the SAW's automatic registration algorithm fails to align the image and gene expression matrix. Possible scenarios include: 1) Incorrect image orientation, i.e., mirrored vs. predefined standard, which can be simply solved by flipping the image [1]; 2) Symmetrical/axis-symmetrical tissue morphology, creating multiple alignment possibilities; 3) Tissue-heatmap discrepancies, leading to erroneous center-of-mass calculations; and other undefined cases.
Manual registration serves to correct registration failures via two options: morphology-based and feature-point. The feature point registration is the preferred method because it delivers quicker and more accurate alignment, particularly for challenging cases such as 2) and 3) mentioned above. It technically only requires one click on the pre-defined dot at the left bottom of the chip to complete the registration. However, its application requires several prerequisites, including newer chip versions (including Stereo-seq N FFPE V1.0, Stereo-seq T FF V1.3, or Stereo-CITE T V1.1), and a passed Image QC, and image in the proper orientation (with the chip handle positioned on the right side).
If feature point registration isn’t available, users should default to the more universal morphology-based option. This method relies on manual adjustment tools and visual verification of alignment results, making it more labor-intensive. Nevertheless, it provides complete control through transformation tools including rotation, flipping, scaling, step moving, and translation to precisely align microscopy images with gene expression heatmaps. Complete documentation for both methods can be found in StereoMap User Manual: [Feature Point Registration] and [Morphology Registration].
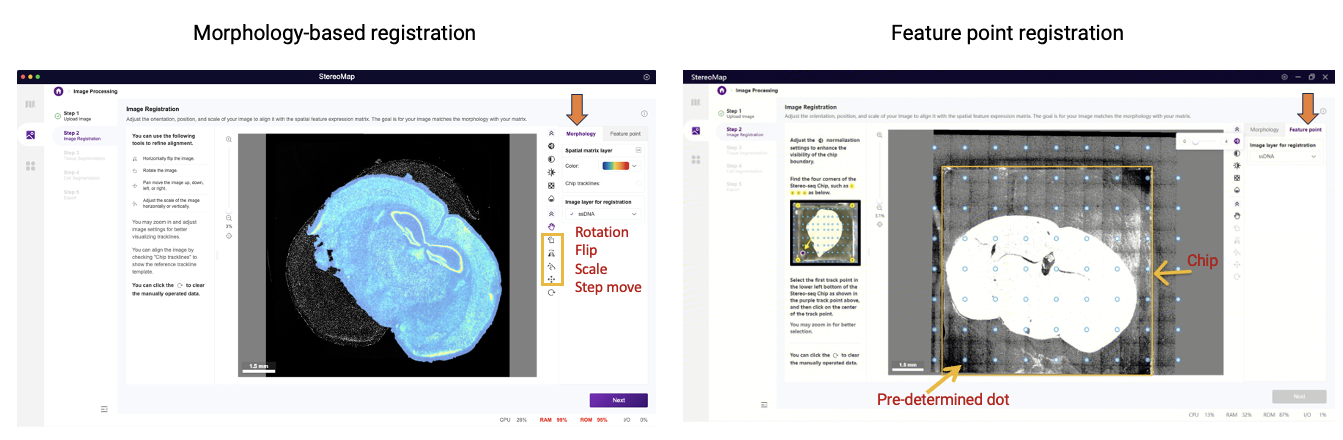 Figure 2. Manual registration options in the StereoMap Image Processing module. From StereoMap User Manual.
Figure 2. Manual registration options in the StereoMap Image Processing module. From StereoMap User Manual.
Tissue segmentation errors may occur after successful registration, manifesting as incomplete tissue coverage or inclusion of non-target regions. While SAW's algorithm often over-segments at tissue edges or when tissue has a complicated discontinuous structure, StereoMap enables rapid corrections using its interactive tools, including brush, eraser, and lasso. For more extensive modifications—especially when the default segmentation poorly matches the sample type—third-party tools like QuPath can generate new masks for import into StereoMap.
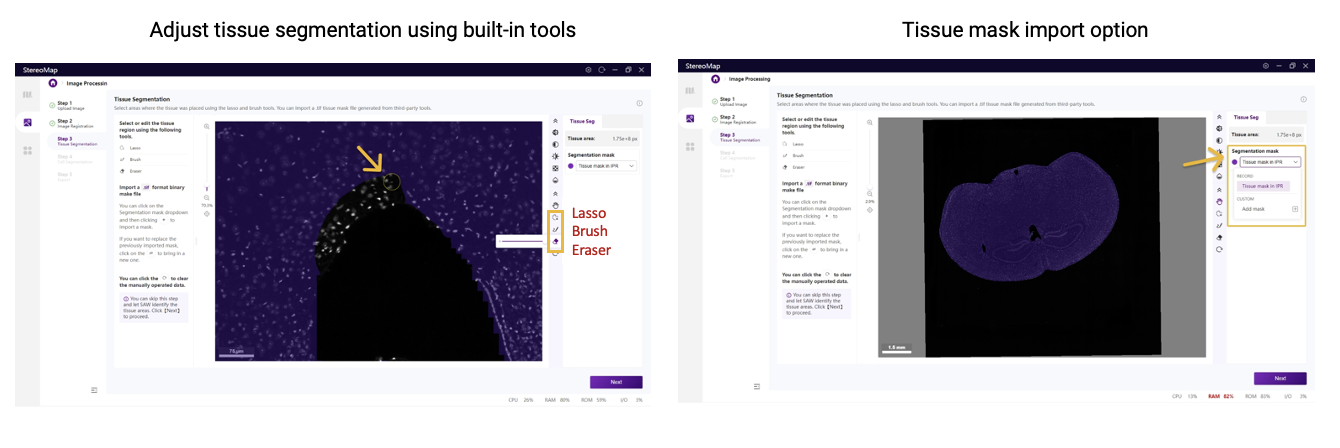 Figure 3. Correcting tissue segmentation errors in StereoMap. From StereoMap User Manual.
Figure 3. Correcting tissue segmentation errors in StereoMap. From StereoMap User Manual.
Cell segmentation errors reflect discrepancies between segmented cells and actual cell morphology. For small errors (local missing or duplicate cells), corrections can be made in StereoMap using interactive editing tools. For major errors, recreate segmentations using third-party tools, such as deep learning-based software Cellpose and DeepCell, and classic thresholding-based method like QuPath. The new cell mask is then imported into StereoMap. Users can choose the appropriate tool based on performance on their specific sample.
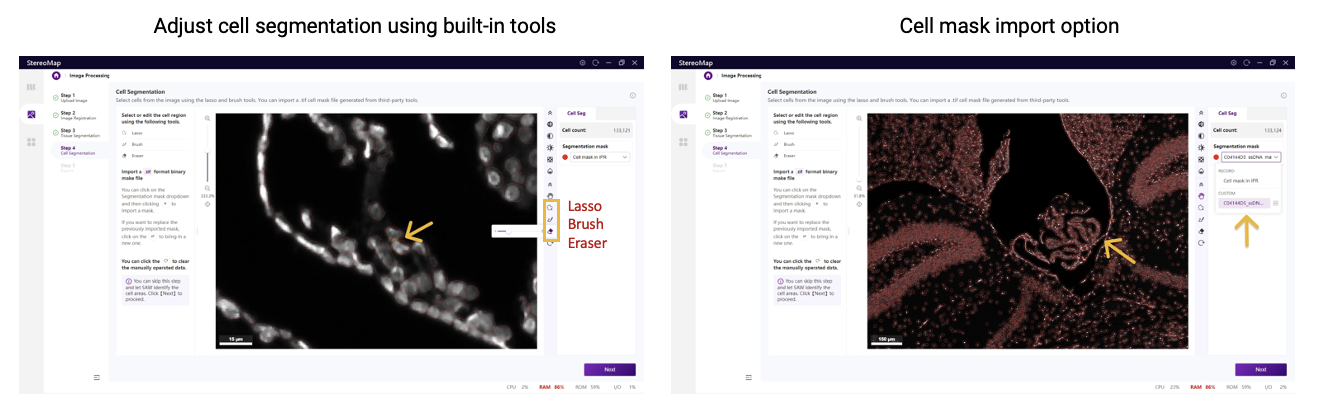 Figure 4. Resolving cell segmentation errors in StereoMap. From StereoMap User Manual.
Figure 4. Resolving cell segmentation errors in StereoMap. From StereoMap User Manual.
For all image processing issues, StereoMap is the central platform for manual corrections. Its intuitive interface enables direct adjustments while supporting imported segmentations from external tools. After completing corrections at the target processing step, users should export the modified image as a new <image.tar.gz> file and resume analysis through the SAW realign pipeline. Below is the command syntax compared to SAW count. For more detailed coding instructions, refer to [SAW User Manual-With manually processed files].

Figure 5. SAW count and realign pipeline commands. The realign pipeline integrates results from the previous SAW count run and the adjusted image file exported from StereoMap.
1. This blog offers a structured troubleshooting table for image processing issues, helping researchers efficiently identify and resolve problems.
2. StereoMap is the central platform for manual image corrections and third-party segmentation imports.
3. SAW realign pipeline integrates with StereoMap's manual image processing results to resume analysis after manual adjustments.
4. Failed Image QC: Run SAW count ("no image" mode) → manual registration → SAW realign.
5. Registration errors: Manual registration offers morphology-based (universal) and feature-point (easier but chip-dependent) options.
6. Tissue/cell segmentation errors: For local refinements, use StereoMap's built-in editing tools. For large-scale adjustments, import corrected masks from third-party tools.
1. Blog | Stereo-seq Image Processing: Essential Steps and their key points. https://en.stomics.tech/news/stomics-blog/1123.html
2. StereoMap User Manual: https://en.stomics.tech/service/stereoMap-operation-manual.html
3. SAW User Manual: https://en.stomics.tech/service/new-saw-operation-manual.html
4. ImageJ website: https://imagej.net/ij/
5. QuPath website: https://qupath.github.io/
6. Cellpose website: https://cellpose.readthedocs.io/en/latest/
7. DeepCell website: https://deepcell.readthedocs.io/en/master/index.html