All
Products
Resources
News
FAQ
Search
21/01/2025
Spatially resolved transcriptomics, named "Method of the Year 2020" by Nature Methods, represents a groundbreaking technology poised to revolutionize the field of genomics by seamlessly integrating transcriptome data with spatial context. It has been broadly used in multiple areas such as elucidating organ organization and function, deciphering the mechanisms of disease onset and progression, unlocking the biological dynamics during development, and understanding evolution at spatial resolution. STOmics' Stereo-seq (SpaTial Enhanced REsolution Omics-sequencing) technology leads in technological advancement, providing researchers with an advanced tool for exploring cellular genomics. STOmics offers true single-cell resolution and unprecedented centimeter-level field-of-view spatial multi-omics capabilities, allowing for simultaneous study and analysis of the multi-omics information at both tissue and cellular levels using fresh frozen and Formalin-Fixed Paraffin-Embedded tissues.
Stereo-seq is a sequencing-based spatial multi-omics platform that is not limited by pre-designed probes or panels and can capture the whole transcriptome for scientific discovery.
Stereo-seq enables the in-situ capturing of RNA information within tissue and subsequently restores spatial cellular location information through spatial coordination barcoding (Coordinate ID, CID). This advanced approach enables the visualization of spatial gene expression profiles in tissues after sequencing on the DNBSEQ platform, facilitating the development of robust research frameworks to explore the intricate interplay between gene expression, cell morphology, and cellular microenvironments.
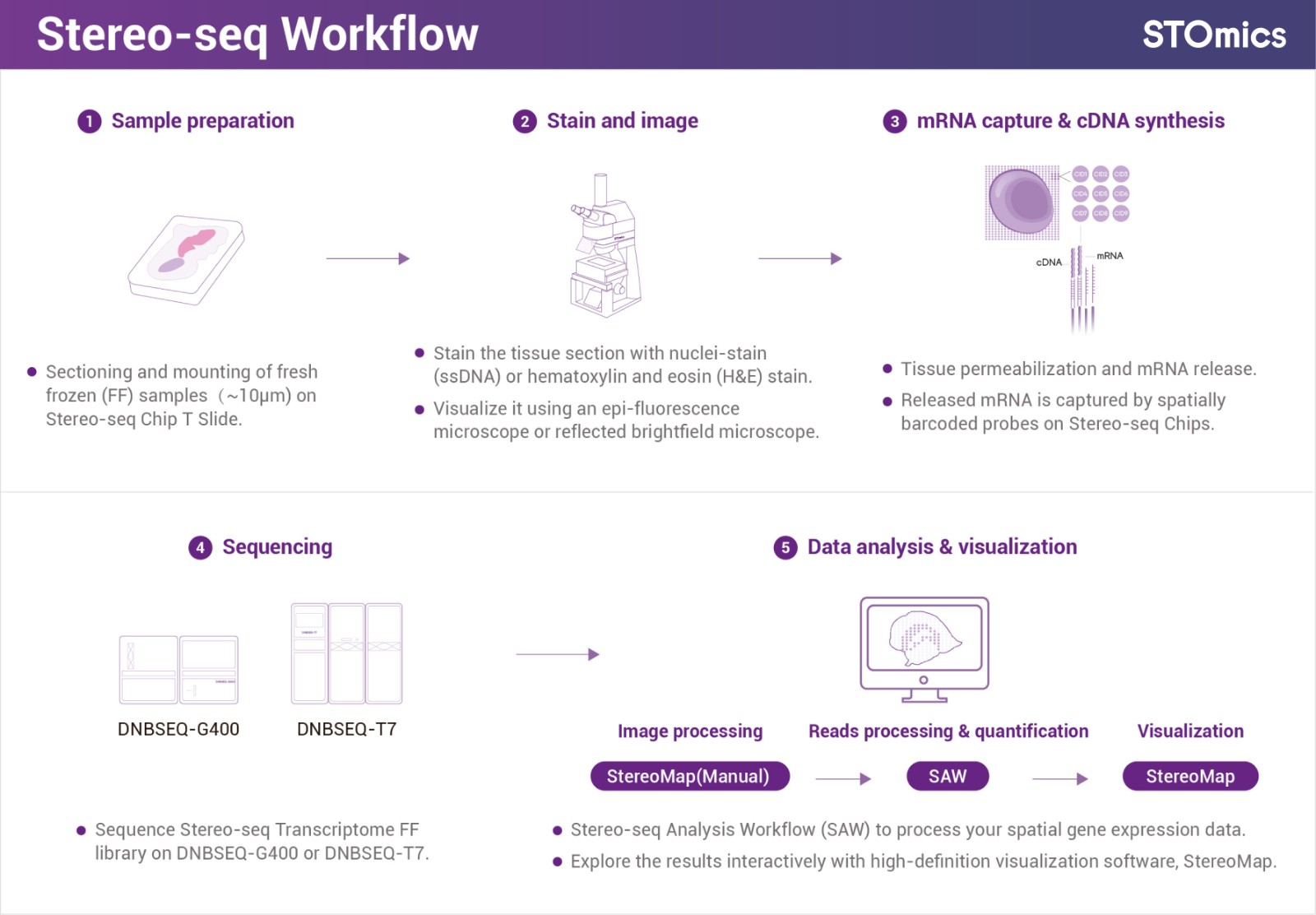
Figure 1. Stereo-seq workflow (Fresh Frozen)
Stereo-seq technology utilizes DNB patterned array chips. The silicon-based Stereo-seq chip features densely patterned RNA-capturing spots, each spot labeled with a unique spatial coordinate barcode (Coordinate ID, CID). A center-to-center distance of just 0.5 µm between adjacent spots enables transcriptome studies and analysis at true single-cell resolution.
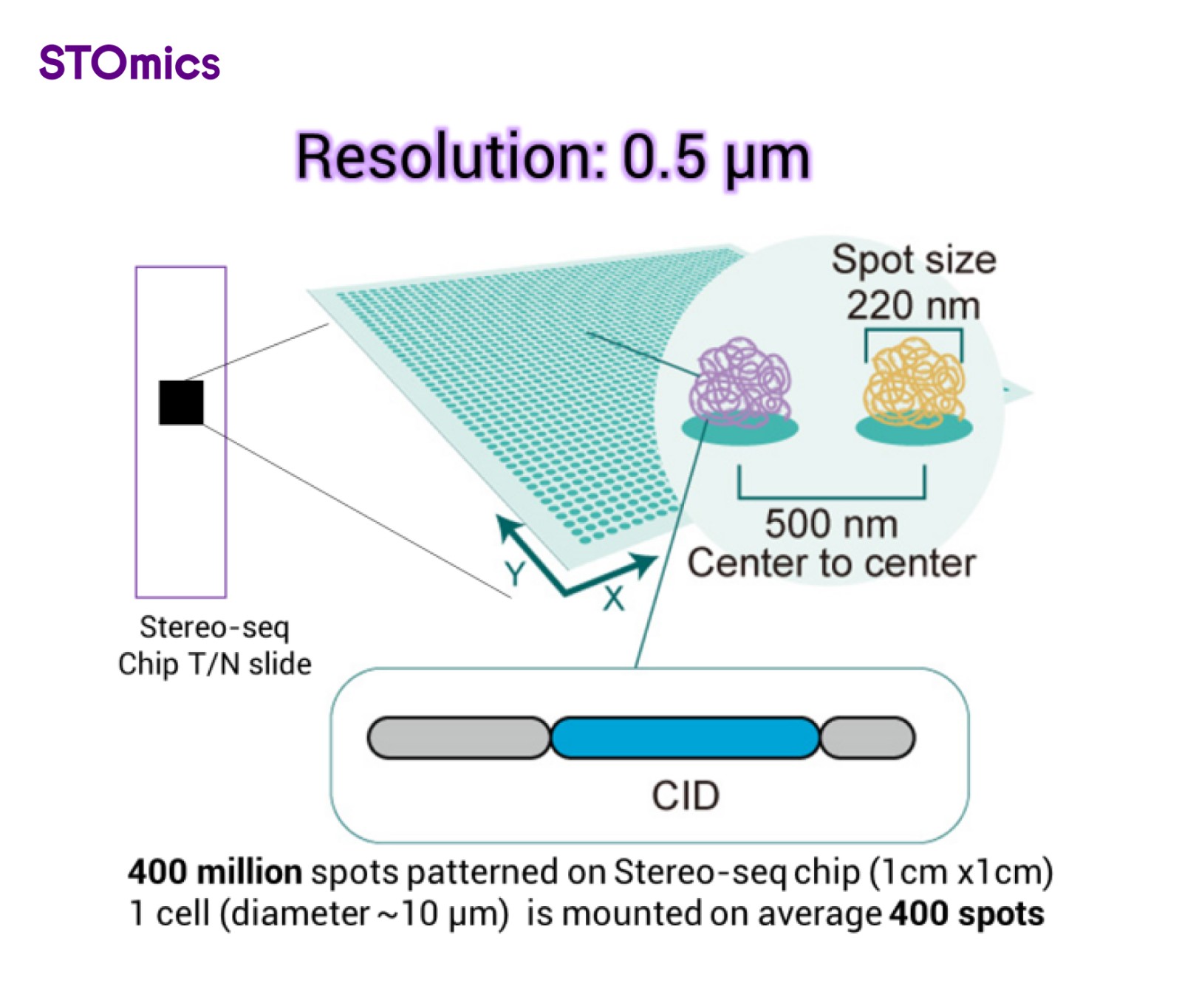
Figure 2. Stereo-seq chip
The standard chip size for Stereo-seq is 1cm x 1cm and can be customized up to 13cm x 13cm. Expanding the capture area allows for profiling of whole mammalian embryos, human organs, or model organisms.
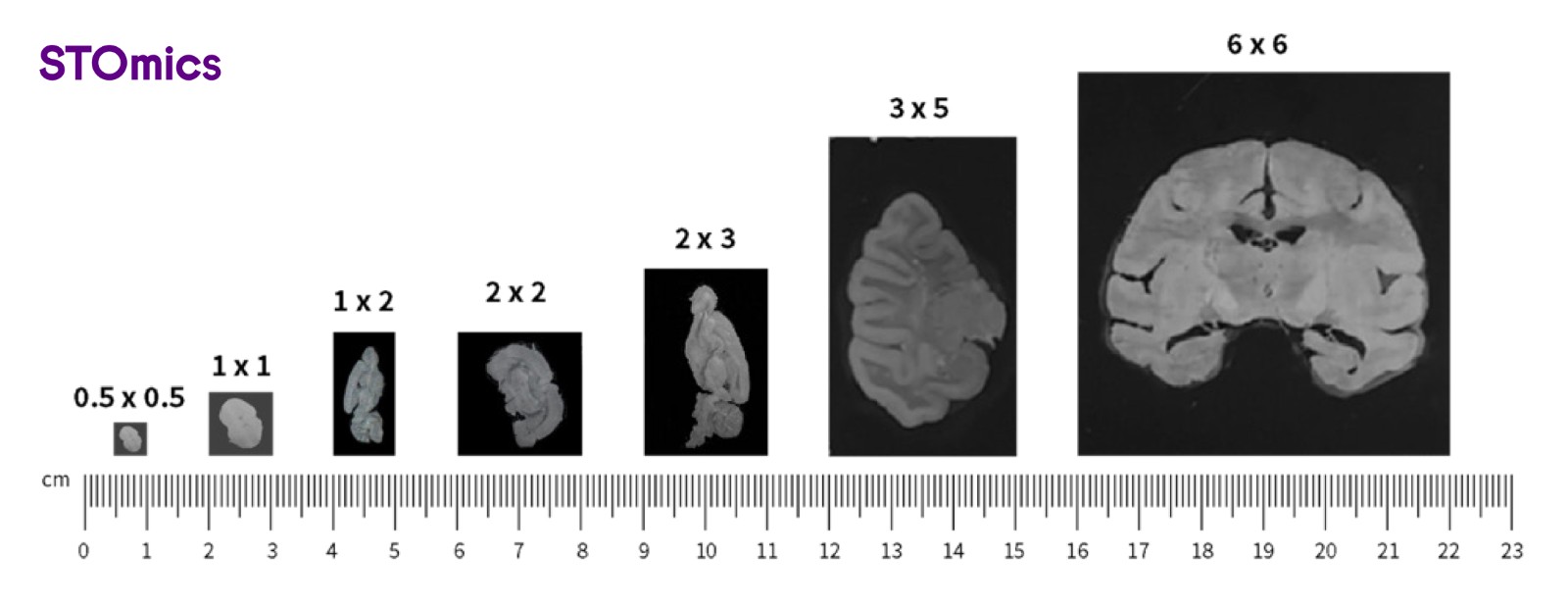
Figure 3. Different Stereo-seq chip sizes
As we dive deep into the spatial world, scientists often demand higher sensitivity and precision. STOmics now presents the upgraded Stereo-seq Transcriptomics Solution v1.3, featuring refined reagent chemistry, enhanced probe design and optimized enzyme selection. This version delivers enhanced capture efficiency, broader compatibility, and a more streamlined workflow while minimizing the diffusion of mRNA molecules to achieve accurate spatial single-cell level analysis.
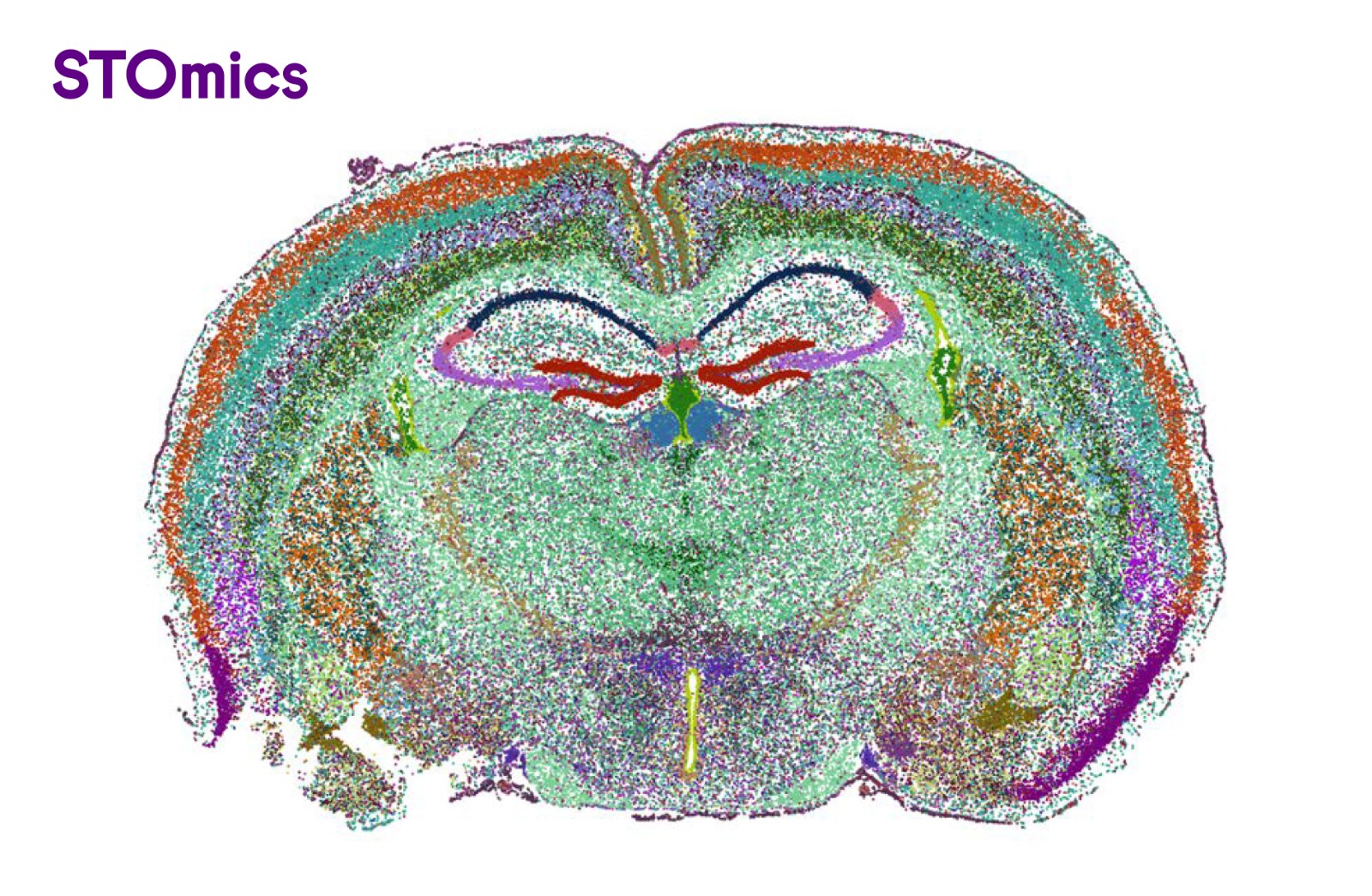
Figure 4. Mouse brain single-cell clustering result (cellbin). 144,000+ cells annotated, 900+ gene type counts per cell, 87 cellular subtypes
The Stereo-seq v1.3 workflow is also compatible with H&E stained tissues. By leveraging the tissue morphological information, Stereo-seq v1.3 assists in identifying specific tissue types, obtaining gene expression profile from specific tissue regions, and performing downstream differential analysis between selected regions of interest.
Spatial transcriptome technologies that utilize Poly-T tailed probes to capture RNA face challenges when applied to FFPE samples, where much of the RNA is incomplete and degraded. Additionally, the probe-based method is limited by predetermined targets or throughput.
The Stereo-seq OMNI for FFPE solution introduces an innovative 'random probe' design that captures total RNA — covering coding RNA, non-coding RNA, and microbial RNA — making it a powerful tool for scientific discovery across various species.
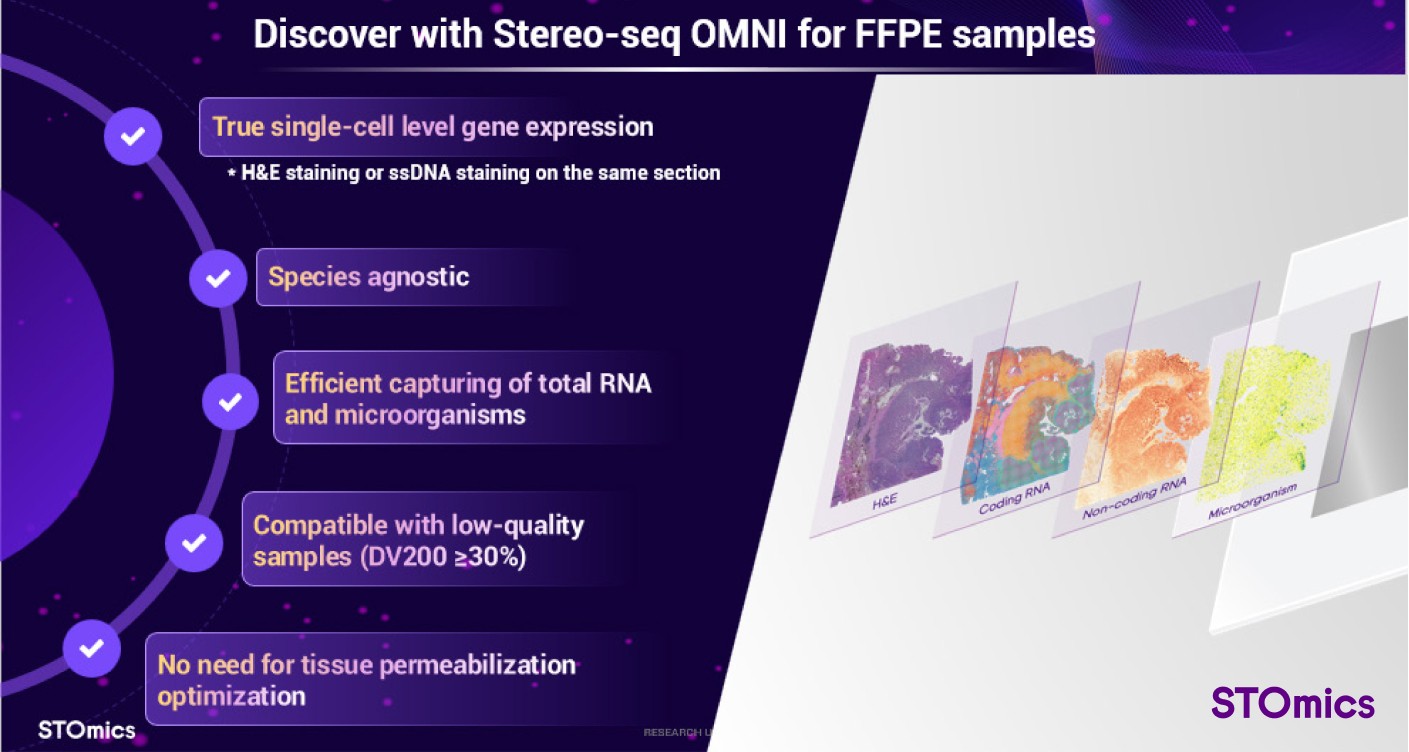
Figure 5. Product highlights of Stereo-seq OMNI
Stereo-seq OMNI enables single-cell analysis using either nucleus or H&E staining on the same tissue section, offering versatile options for different research needs. Additionally, it supports samples with low RNA integrity (DV200≥30%), expanding compatibility for various specimens. The Stereo-seq OMNI workflow does not require tissue permeabilization optimization, which reduces experiment duration and simplifies the overall process.
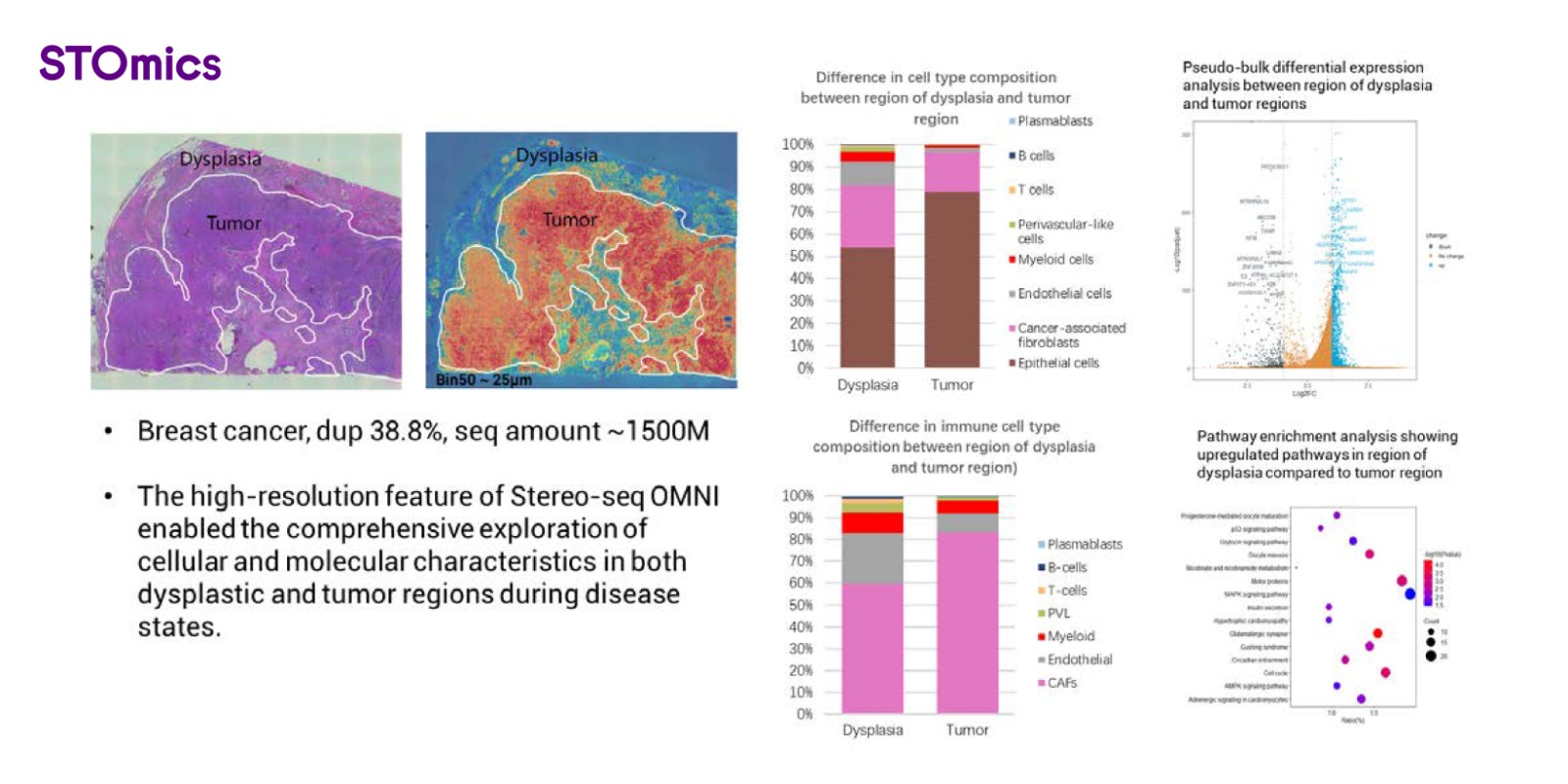
Figure 6. Spatial gene expression profile on breast cancer FFPE sample using Stereo-seq OMNI
Stereo-seq OMNI provides multiple chip size options. The current standard chip size is 1cm x 1cm. Mini chip (0.5cm x 0.5cm) and large chips (1cm×2cm, 2cm x 2cm, 3cm x 2cm) will be available soon to accommodate broader application needs.
Reflecting on the central dogma, biological information flows from DNA to RNA, and then to protein. Studies have shown that RNA and protein information are not entirely aligned, with the correlation of RNA and protein across genes varying significantly among different tissues (5). Consequently, relying solely on single omics data does not provide a comprehensive view; instead multi-omics data can reflect this diversity and address the intricate biological questions pertaining to all processes of birth, aging, illness, and death.
Stereo-CITE integrates the Stereo-seq technique and Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) to detect high-plex proteins with spatial whole transcriptome simultaneously in the same sample. The Stereo-seq Chip T (poly-T-based chip) is loaded with capture probes that contain spatial coordinate information. Through a series of biochemical procedures (Figure x), the probes capture mRNA molecules and antibody-derived tags (ADTs) in situ within the tissue, enabling cDNA synthesis and obtaining transcriptome data alongside multi-protein spatial distribution information of the entire tissue through sequencing and a corresponding visualization platform.
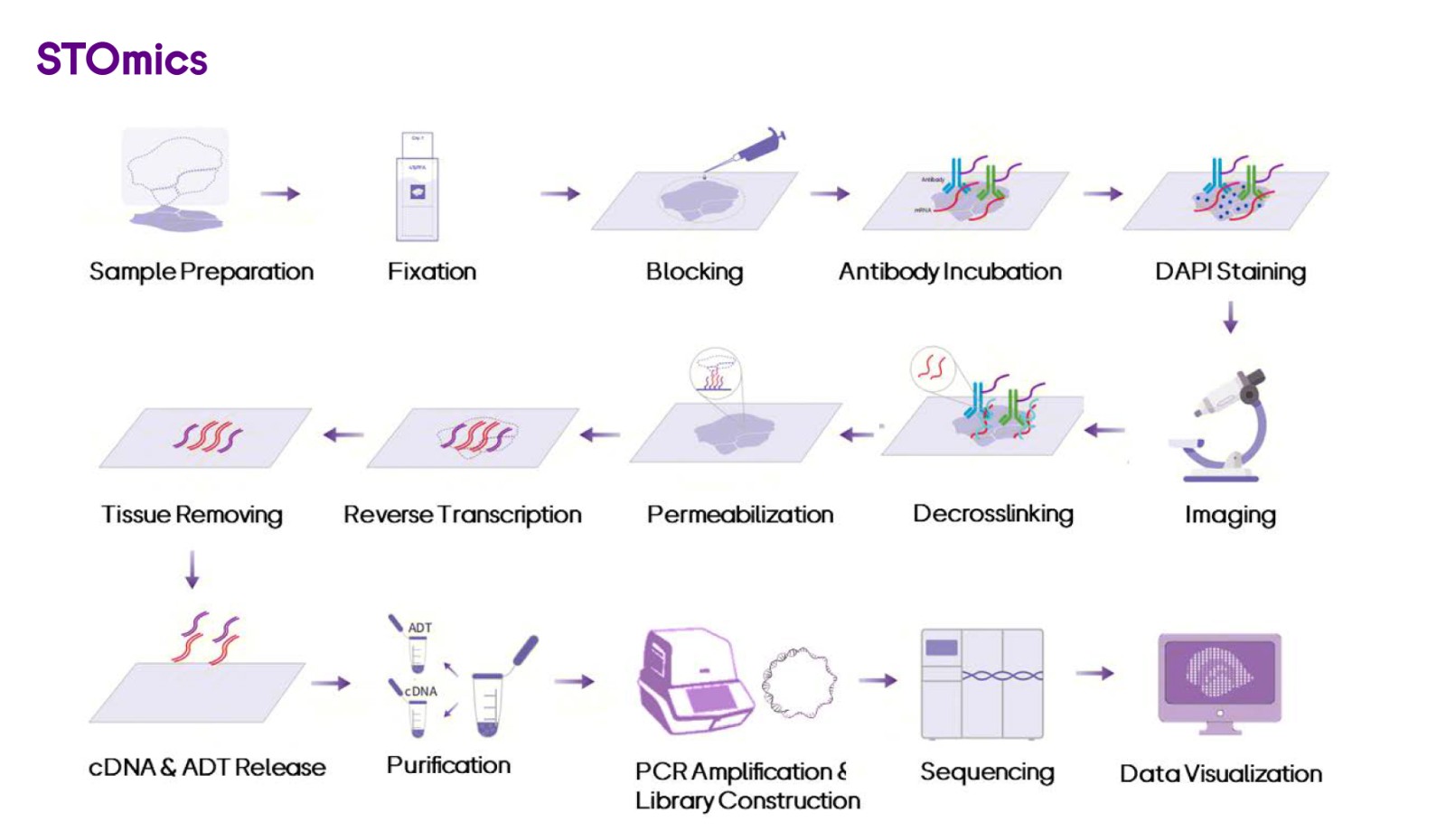
Figure 7. Whole workflow of Stereo-CITE
Stereo-CITE provides single-cell resolution for analyzing both transcriptomes and proteins in the same fresh frozen tissue section. It supports more than 100 plex proteins, utilizing antibody combinations from validated in-house vendors.
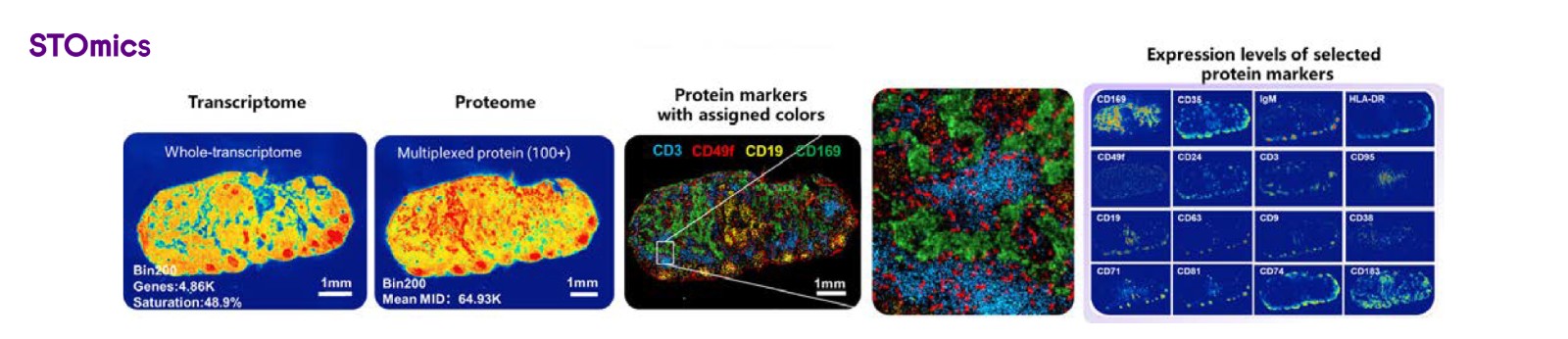
Figure 8. Demo of Stereo-CITE on human paracancerous lymph nodes sample
Stereo-CITE enhances spatial protein profiling precision by preventing autofluorescence interference and antigen instability caused by multiple rounds of testing. It ensures a multi-modal approach to cell phenotyping and the biological mechanisms relevant to tumor heterogeneity, tumor microenvironments, cellular networks, and therapeutic responses.
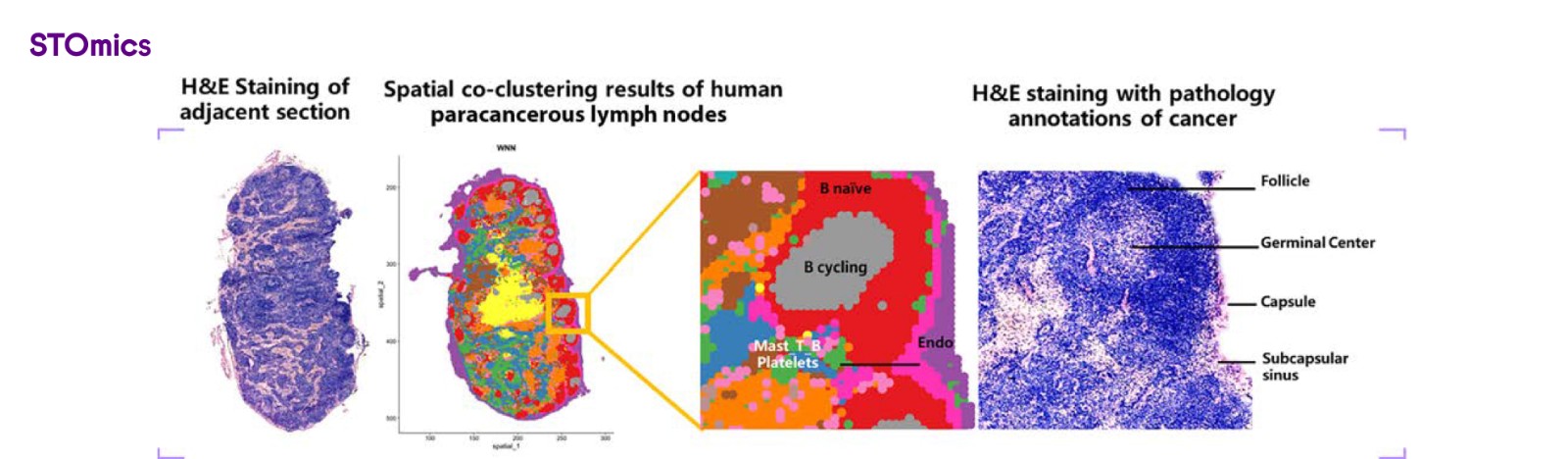
Figure 9. Stereo-CITE recapitulated the organizational structure of human lymph nodes
To fulfill the fast-growing needs of multi-omics applications on clinical FFPE samples, STOmics is developing the next version of the Stereo-CITE total solution, which is expected in the second half of 2025.
STOmics offers a comprehensive solution that encompasses both the kit and reagent products, along with user-friendly software pipelines, Stereo-seq Analysis Workflow (SAW) and StereoMap, and dedicated technology support. Customers can utilize the software pipelines to generate spatial gene expression profiles and visualize their Stereo-seq data. Additionally, the Stereo-seq data is compatible with third-party open-sourced tools, enhancing the analysis capabilities for user's data.
1. Chen A, Liao S, Cheng M, et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays[J]. Cell, 2022, 185(10): 1777-1792. e21.
2. Yue Y, et. al. Systematic comparison of sequencing-based spatial transcriptomic methods. Nature Methods, 2024.
3. Moffitt JR, et al. The emerging landscape of spatial profiling technologies. Nat Rev Genet. 2022 Dec;23(12):741-759. doi: 10.1038/s41576-022-00515-3. Epub 2022 Jul 20. PMID: 35859028.
4. Ou Z., Yin J., Wu L., et al., (2023). Spatial transcriptomics in cancer research: Opportunities and challenges. The Innovation Life 1(1), 100006. https://doi.org/10.59717/j.xinn-life.2023.100006
5. Buccitelli C, et al. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21(10):630-644 (2020).