All
Products
Resources
News
FAQ
Search
25/02/2025
Spatial Transcriptomics (ST) methods supplement the location information lost using scRNA-seq approaches for spatial biology research. Emerging large field-of-view (FOV) ST platforms enable whole-embryo-scale spatiotemporal research at different time points. However, how to do the downstream computational analysis for 3D reconstruction, spatial domain digitization, and cell-cell interaction still blocks the way for further digging embryo-scale and 3D ST dataset.
An international team of scientists from leading research institutes worldwide collaborated to conduct pioneering research in building a 3D spatiotemporal modeling framework—Spateo (https://github.com/aristoteleo/spateo-release). Their accomplishment was published as a featured article in Cell, titled “Spatiotemporal modeling of molecular holograms”.
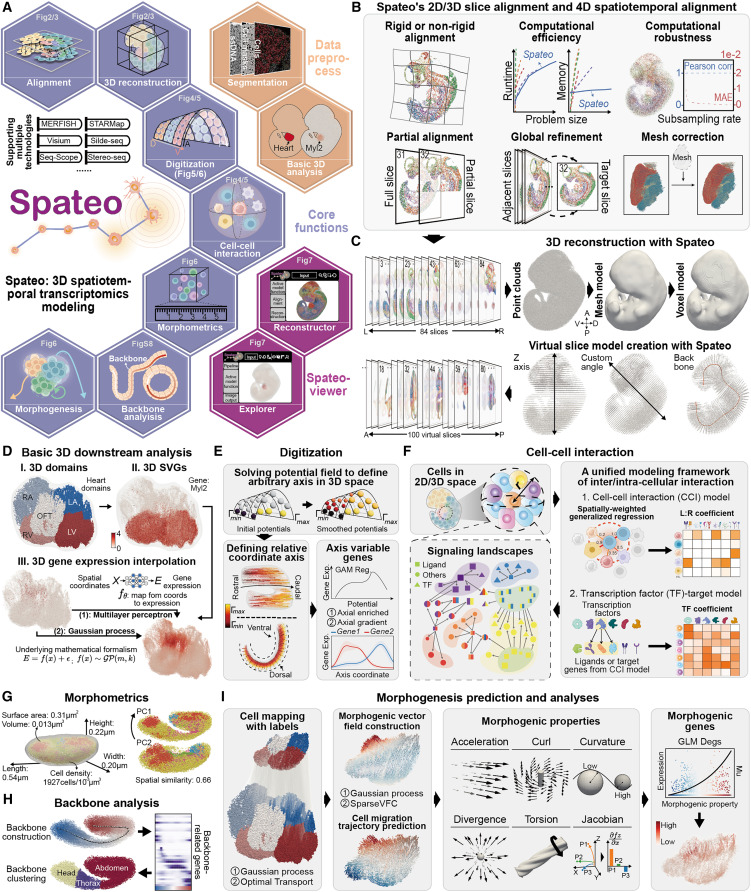
Spateo, a versatile tool for 3D spatiotemporal transcriptomics modeling at single-cell resolution and whole-embryo scale, demonstrated on the mouse and Drosophila embryos.
Understanding how cells migrate and interact during early development is crucial for uncovering the causes of congenital diseases. Current spatial omics tools are great for visualizing spatial expression patterns but lack the ability to track cellular changes over time. Spateo offers a unified framework that allows Answers to create highly detailed 3D "molecular holograms" of whole embryos, making it easier to study dynamic developmental processes.
One of the biggest challenges in reconstructing 3D tissues from sections is the loss of information due to missing regions and tissue deformations. Many existing methods struggle with aligning large-scale embryonic sections, especially when dealing with local distortions. Spateo introduces an advanced mathematical approach using pairwise alignment, multi-slice refinement, and mesh correction, which significantly improves the accuracy of alignment and large-scale tissue reconstruction.
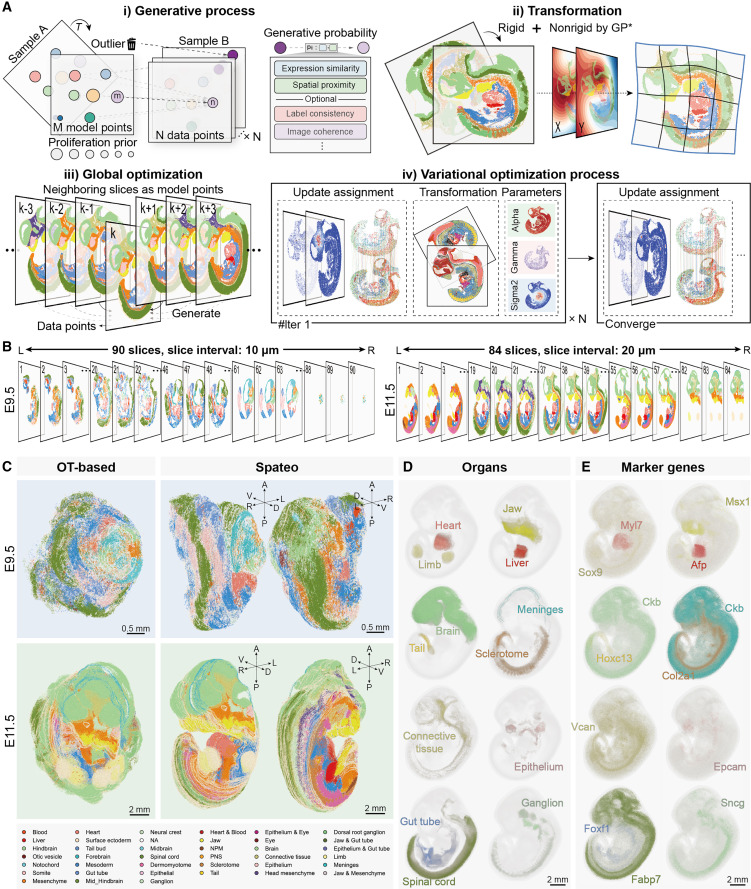
Spateo enables accurate, efficient, and scalable reconstruction of 3D molecular holograms of whole mouse embryos.
Spateo can handle both rigid and non-rigid transformations, allowing it to compensate for distortions and missing sections. Even complex tissues can be reconstructed in 3D with remarkable precision. By applying the algorithm to biological systems like the mouse forebrain hemisphere, human metastatic lymph nodes, macaque cortex, and mouse embryo, we confirmed that Spateo outperforms existing state-of-the-art methods. We also provide an editable visualization tool, Spateo-view, for research and publication purposes.
Embryonic regionalization is how an embryo is divided into different compartments, each forming specific tissues or organs. Spateo introduces a novel digitization method that enables the exploration of gene expression gradients in topologically complex structures. Additionally, it incorporates a spatial regression function to infer upstream and downstream gene regulations through cell-cell interaction (CCI) analysis.
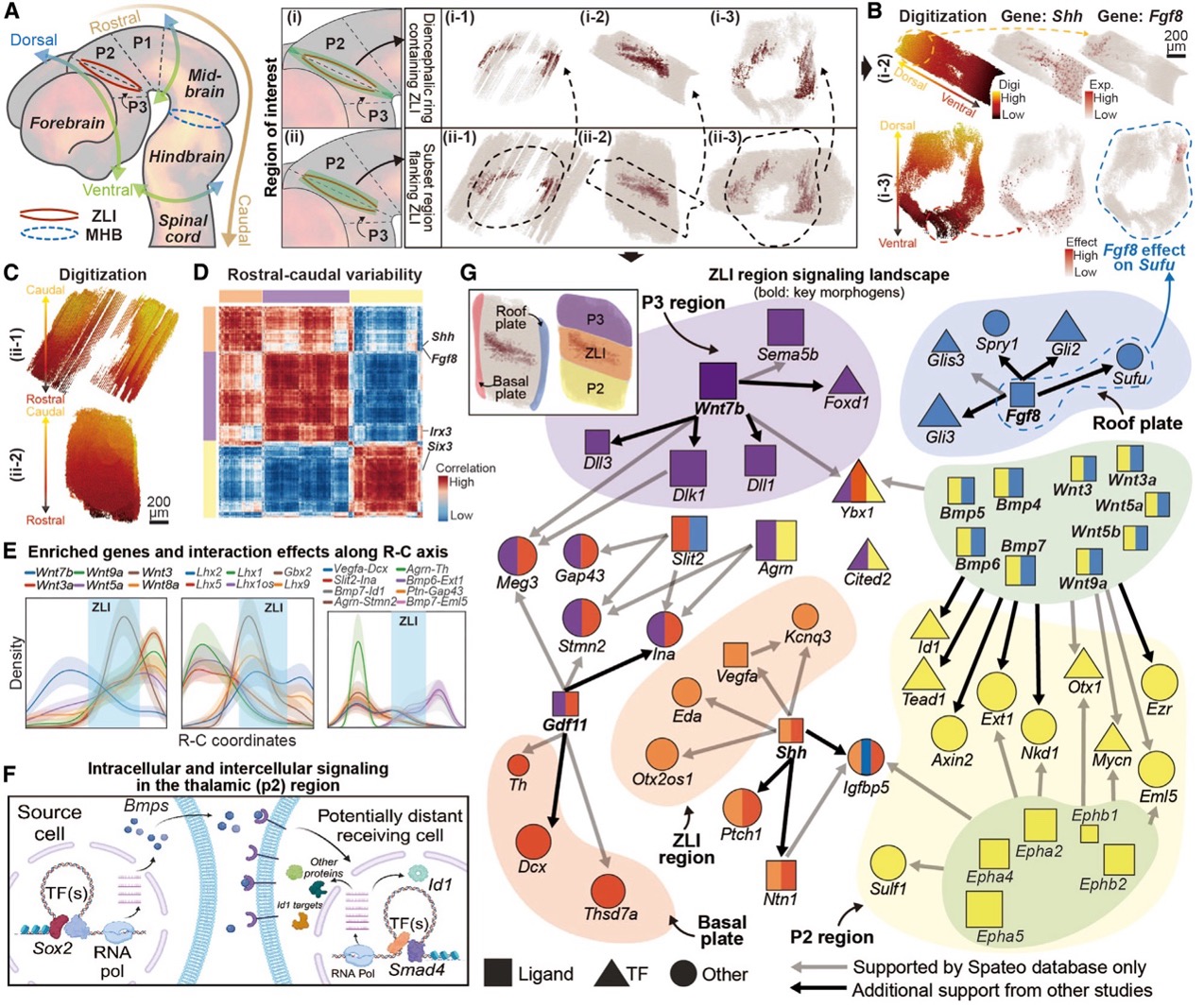
Spateo identifies networks of intercellular signaling and intracellular regulations in the developing brain proximal to the zona limitans intrathalamica.
Spateo was demonstrated on a 3D mouse embryo dataset, enabling us to investigate structures such as the central nervous system (CNS) and the developing heart. These insights provide a clearer picture of how gene expression patterns drive cell fate determination during development. The same methods can be applied to studying diseases such as non-small cell lung cancer and understanding complex brain regions like the macaque cortex.
This is, to our knowledge, the first attempt to link microscopic RNA expression to macroscopic morphogenesis. These vector fields allow us to measure the physical properties of tissue development, including 3D curl (rotation), acceleration, curvature, torsion, divergence (expansion/shrinkage), and the Jacobian matrix (how movement along one axis affects another). These mathematical properties have real biological meaning, helping us uncover key morphogenetic genes responsible for shaping tissues.
A great example is heart development. Spateo uncovered distinct cell movement patterns, showing how the right atrium (RA) exhibited the highest acceleration, while the right ventricle (RV) and left atrium (LA) had significant curl, indicating rotational movements. In contrast, the left ventricle (LV) showed the lowest divergence, suggesting minimal expansion or contraction. These findings align with the developmental timeline of the heart and could provide new insights into congenital heart defects.
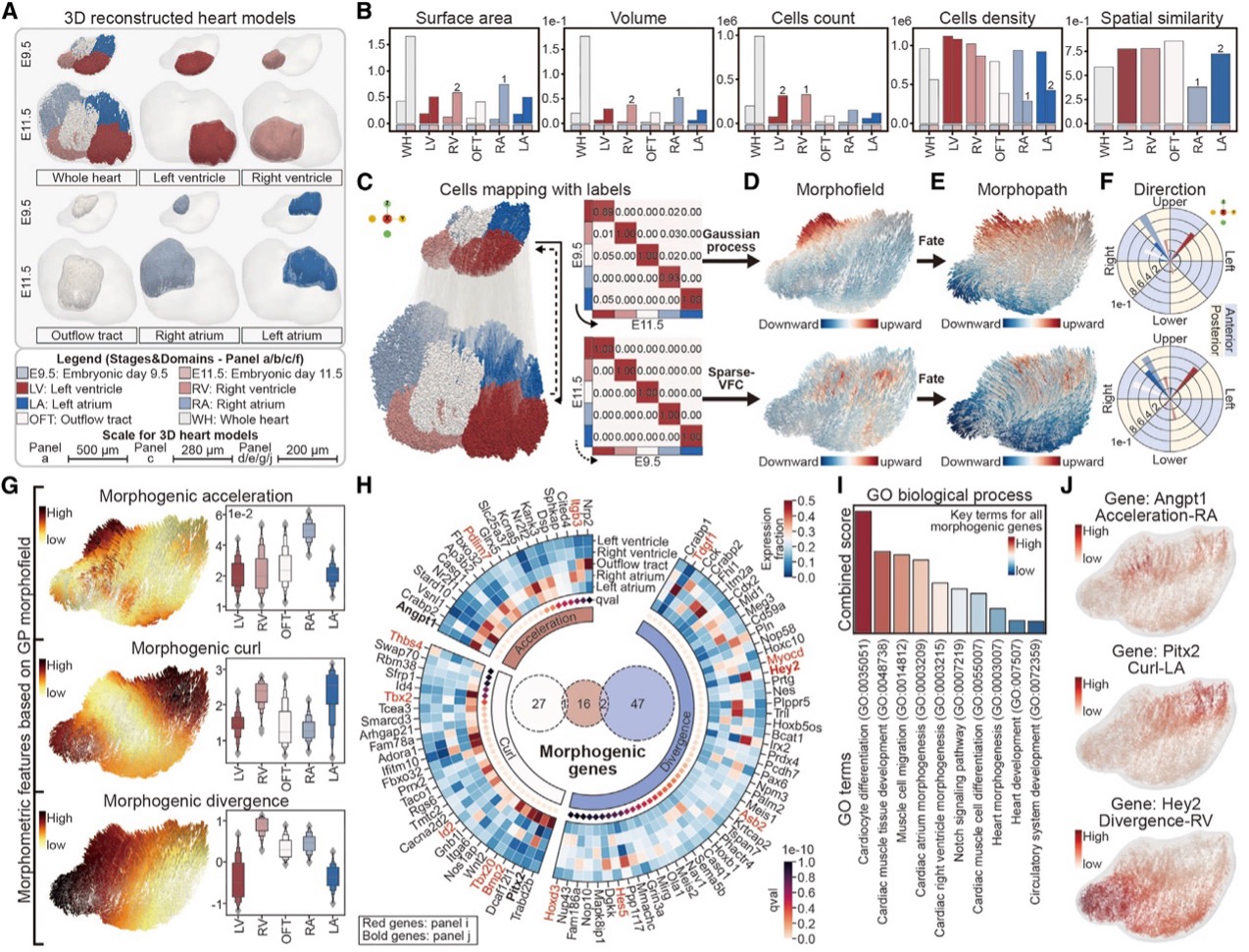
Spateo characterizes morphometric and molecular dynamics involved in the asymmetrical heart organogenesis.
We would like to demonstrate our algorithms on a sequencing platform that offers both a field of view at the whole embryo scale and molecular readability down to single-cell resolution. The Stereo-seq platform was capable of these features at an affordable cost at that time. We, therefore, created the mouse and Drosophila embryonic 3D spatial transcriptomics datasets for this research. These data will be used for further comprehensive studies. The processed data can be downloaded from this webpage (http://spateodata.aristoteleo.com/).
As spatial transcriptomics technology continues to evolve, Spateo will be instrumental in cross-species comparisons of organ development, potentially revealing evolutionary patterns. It could also be applied to studying complex 3D tissues in human diseases. As spatial technologies mature, single-cell genomics techniques will increasingly be adapted for spatial genomics, enabling more profound studies of cells in their natural 3D environments.
Learn more about the research: https://www.cell.com/cell/fulltext/S0092-8674(24)01159-0
More reports about this research: Nature https://www.nature.com/articles/d41586-024-03615-8
Nature methods https://www.nature.com/articles/s41592-024-02587-x