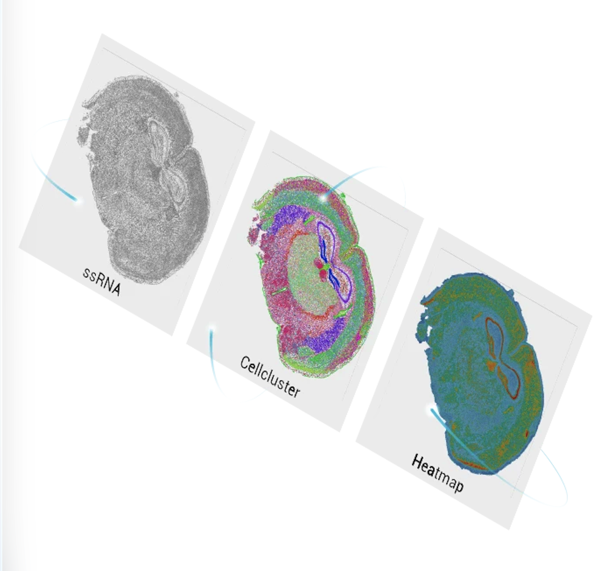
To understand how the brain works, researchers need to connect molecular identity with spatial organization - from single cells to entire neural circuits. Stereo-seq enables this connection by providing single-cell resolution transcriptomics across large brain areas, allowing genes, cells, and circuits to be studied within their native spatial context.
Why Spatial Context Matters in Neuroscience
The brain’s layered structure, cellular diversity, and long-range connectivity mean that gene expression must be interpreted within its native spatial environment. Understanding how molecular programs vary across regions and circuits therefore requires tools that preserve tissue organization while maintaining molecular resolution.
The Limitations of Current Tools
While single-cell RNA sequencing has transformed cell-type classification, tissue dissociation eliminates spatial relationships (Lee et al., 2022). Conventional spatial transcriptomic methods retain tissue structure but often sacrifice resolution, transcriptome depth, or scalability (Park et al., 2023). Together, these limitations force a trade-off between molecular detail and spatial context, limiting interpretation at the level of neural circuits and whole brain regions.
How Stereo-seq Overcomes the Resolution-Coverage Trade-off
Stereo-seq was developed in direct response to this trade-off. By combining single-cell resolution transcriptomic profiling with large-area spatial coverage, it eliminates the need to choose between molecular detail and intact tissue architecture. Using ultra-high-density DNA NanoBall (DNB) arrays with a 500 nm center-to-center spacing, Stereo-seq captures whole-transcriptome information at single-cell resolution (Fig. 1). Hundreds of mRNA molecules can be detected within a single cell, each assigned precise spatial coordinates, enabling:
Identify cell types robustly
Resolve spatial heterogeneity of transcripts within individual cells
Study neuronal polarity, synapses, and single-cell disease changes
 arranged at single-cell resolution, enabling mRNA from individual cells to be detected with hundreds of DNBs, each carrying a unique coordinate ID._202601071725.png)
Figure 1. Stereo-seq captures DNA nanoballs (DNBs) arranged at single-cell resolution, enabling mRNA from individual cells to be detected with hundreds of DNBs, each carrying a unique coordinate ID.
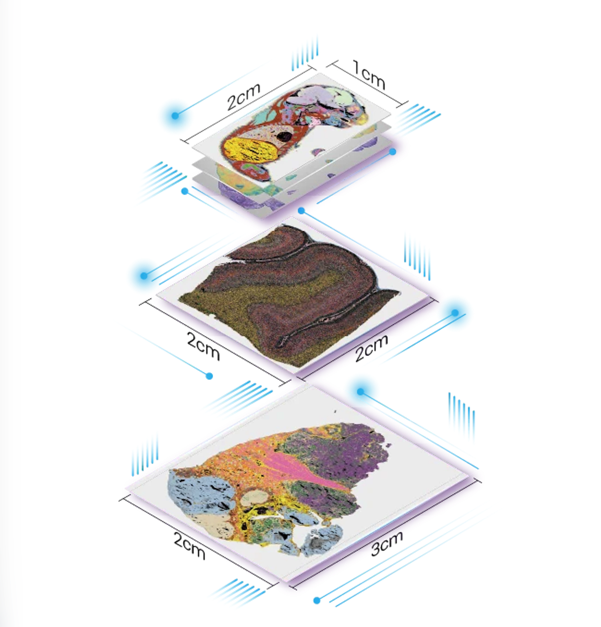
Stereo-seq is also scalable and customizable. It offers multiple chip sizes, with custom arrays available up to 13 cm × 13 cm, enabling spatial transcriptomic analysis from localized brain regions to entire brain sections (Fig. 2). This means molecular details at the cellular level can be directly linked to large-scale brain architecture - all in a single experiment.
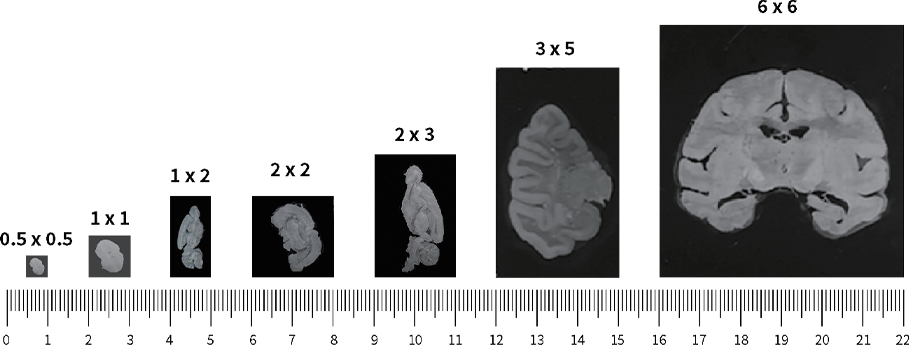
Figure 2. Demonstration of Stereo-seq chips of different sizes.
Stereo-seq in Action: New Insights into the Brain
Stereo-seq has already been applied to a variety of challenging questions, from mapping elusive brain regions to understanding neurodegenerative diseases and cross-species comparisons.
Mapping the macaque claustrum in spatial context
The claustrum is one of the most enigmatic brain regions, in part due to its thin morphology and widespread connectivity. To overcome these challenges, Lei et al. (2025) used Stereo-seq Transcriptomics FF Solution (5cm x 3cm, 1cm x 2cm, and 1cm x 1cm) to generate the first single-cell spatial transcriptomic atlas and whole-brain connectivity map of the macaque claustrum. Their analysis identified 48 distinct cell types and revealed extensive connections with both cortical and subcortical regions, providing new insight into the organization and potential function of this elusive structure.
Resolving Alzheimer’s disease pathology at single-cell resolution
Gong et al. (2025) applied Stereo-seq Transcriptomics FF Solution (1cm x 1cm) to generate single-cell resolution transcriptomic maps of the human prefrontal cortex in Alzheimer’s disease. Their study revealed disease-associated transcriptional alterations, disrupted cortical layering, and changes in cell-cell communication - features that would be difficult to detect without preserving native tissue organization.
Comparative spatial transcriptomics across species
Hao et al. (2024) leveraged Stereo-seq Large Chip Design (LCD)/Customized LCD to map the cerebellar cortex of macaques, marmosets, and mice, uncovering primate-specific cell types and spatial gene expression patterns. This comparative framework linked molecular organization to functional connectivity and evolutionary divergence across species.
These studies show how Stereo-seq bridges the gap between molecules, cells, and circuits.
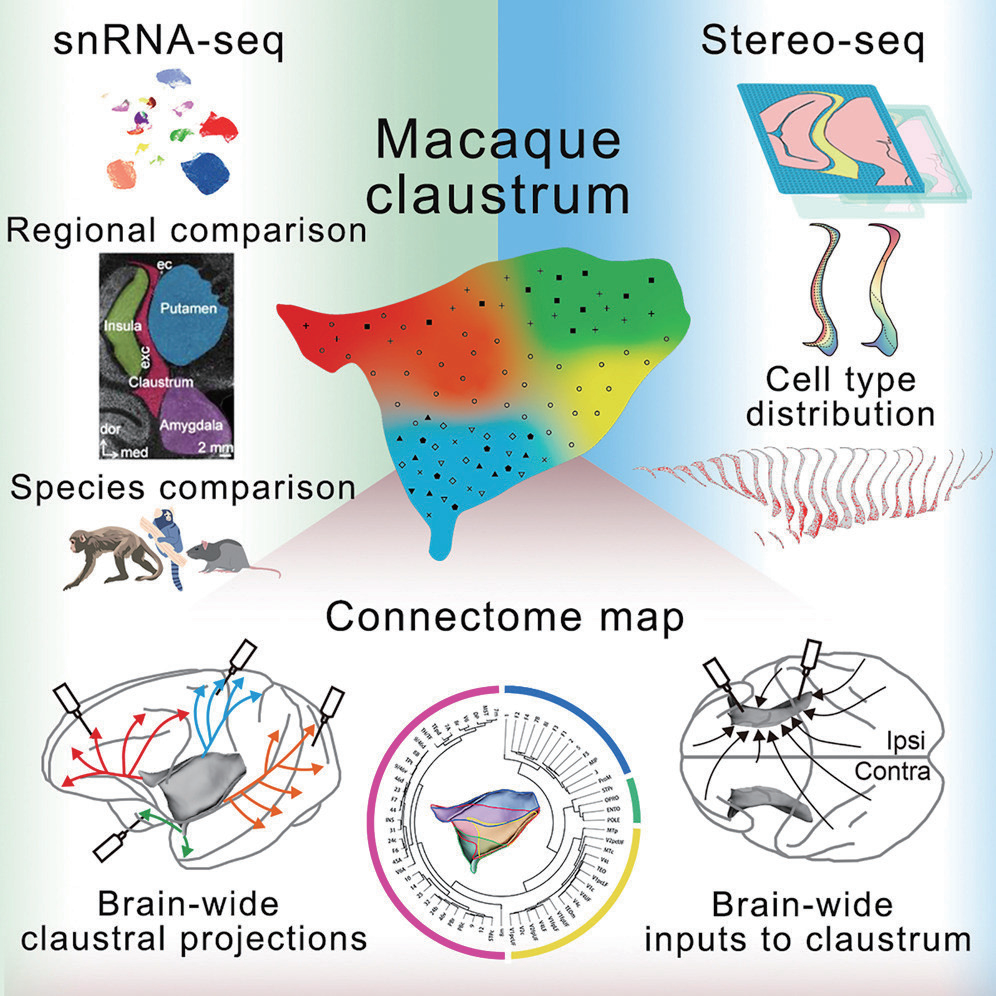
Image from Lei et al., Cell, 2025, licensed under CC BY 4.0
A Spatial Blueprint for Next-Generation Neuroscience
Stereo-seq preserves native tissue structure at single-cell resolution while covering large areas of the brain, providing a unified framework to connect genes, cells, and neural circuits. For studies of brain development, circuit organization, and neurological disease, this capability enables molecular mechanisms to be interpreted directly within their anatomical and functional context. As spatial neuroscience continues to evolve, Stereo-seq offers a foundation for exploring the brain across scales.
STOmics Product Matrix: Scalable Solutions Across Spatial Scales
STOmics leverages core Stereo-seq technology to provide scalable spatial transcriptomics solutions. The following product matrix summarizes the available options and their typical use cases (Table 1). All products share the same ultra-high-density DNA NanoBall (DNB) array architecture, ensuring consistent single-cell resolution and transcriptome depth across different spatial scales.
Table 1. STOmics product matrix
This product matrix allows researchers to move seamlessly from localized circuits to whole-brain systems while preserving molecular resolution, spatial context, and transcriptome depth. If your research requires tailored spatial transcriptomics solutions or support in experimental design, feel free to Contact Us or reach out directly at info_global@stomics.tech. Our team is ready to support your research goals.